Dionex 063000 User manual

DNAPac PA200 Document No. 065036 Page 1 of 25
PRODUCT MANUAL
DNAPAC PA200 ANALYTICAL COLUMN
(4 x 250mm, P/N 063000)
DNAPAC PA200 GUARD COLUMN
(4 x 50mm, P/N 062998)
©DIONEX Corporation
Document No. 065036
Revision 01
20 August 2004

DNAPac PA200 Document No. 065036 Page 2 of 25
TABLE OF CONTENTS
SECTION 1 - INTRODUCTION ...................................................................................................................4
1.1
DNAPAC PA200 ................................................................................................................................ 4
1.2
BIOLC SYSTEM (WITHOUT COLUMNS) .............................................................................................. 4
1.3
GUARD COLUMN USE ........................................................................................................................ 5
1.4
DNAPAC PA200 ANION EXCHANGE COLUMNS ............................................................................... 5
1.5
DNAPAC COLUMN FAMILY ............................................................................................................... 5
SECTION 2 - OPERATION AND SYSTEM REQUIREMENTS .............................................................. 6
2.1
SYSTEM REQUIREMENTS.................................................................................................................... 6
2.2
SYSTEM OPERATION REQUIREMENTS ................................................................................................ 6
2.3
DNAPAC PA200 COLUMN OPERATIONAL PARAMETERS................................................................. 7
SECTION 3 - PURITY REQUIREMENTS FOR CHEMICALS...............................................................8
3.1
DEIONIZED WATER ............................................................................................................................ 8
3.2
INORGANIC CHEMICALS..................................................................................................................... 8
3.3
SOLVENTS .......................................................................................................................................... 8
SECTION 4 - QUALITY ASSURANCE.......................................................................................................9
4.1
CERTIFICATE OF PERFORMANCE –RESIN BATCH TESTING ............................................................... 9
4.2
PRODUCTION TEST CHROMATOGRAMS ........................................................................................... 10
SECTION 5 - METHODS DEVELOPMENT.............................................................................................11
5.1
SAMPLE CLEANUP ............................................................................................................................ 11
5.2
ELUTION ORDER............................................................................................................................... 11
5.3
EFFECT OF SALT TYPE ON OLIGONUCLEOTIDE ELUTION ................................................................ 11
5.3.1
Eluent Strength ...................................................................................................................... 11
5.3.2
Loading Capacity................................................................................................................... 11
5.4
GRADIENT SLOPE.............................................................................................................................. 12
5.5
EFFECT OF PHAND SOLVENT ON OLIGONUCLEOTIDE CHROMATOGRAPHY .................................... 12
5.5.1
Effect of pH on Hydrogen Bond Interactions ........................................................................ 12
5.5.2 Effect of pH on Retention...................................................................................................... 13
5.5.2
Effect of pH on Retention...................................................................................................... 13
5.5.3
Effect of Solvent on Retention............................................................................................... 13
5.5.4
Effect of pH on Selectivity .................................................................................................... 14
5.6
EFFECT OF TEMPERATURE ON OLIGONUCLEOTIDE RETENTION ...................................................... 15
5.7
EFFECT OF TERMINAL BASE ON SELECTIVITY ................................................................................. 16
5.7.1
Selectivity in Sodium Chloride (NaCl) Gradients. ................................................................ 16
5.7.2
Selectivity in Sodium Perchlorate (NaClO
4
) Gradients ......................................................... 17
5.8
APPLICATION-SPECIFIC MOBILE PHASE RECOMMENDATIONS ........................................................ 18
5.8.1
For synthetic ODNs where the goal is to evaluate purity. ..................................................... 18
5.8.2
When multiple possible ODNs of similar length in the same solution must be resolved. ..... 18
SECTION 6 - APPLICATIONS................................................................................................................... 19
6.1
DENATURING CONDITIONS FOR CONTROL OF SECONDARY STRUCTURE ........................................ 19
6.2
EFFECT OF HIGH TEMPERATURE AND HIGH PHON COLUMN LIFETIME.......................................... 19
6.3
PHOSPHODIESTER ANALYSIS ........................................................................................................... 20
6.3.1
Sodium Perchlorate Eluent Systems ...................................................................................... 20
6.3.2
Sodium Chloride Eluent Systems .......................................................................................... 21
SECTION 7 - DNAPac™ PA200 RESOURCES.........................................................................................22

DNAPac PA200 Document No. 065036 Page 3 of 25
Section 8 - TROUBLESHOOTING GUIDE ...............................................................................................23
8.1
FINDING THE SOURCE OF HIGH SYSTEM BACK PRESSURE .............................................................. 23
8.2
BACKPRESSURE ON COLUMN HAS INCREASED................................................................................ 23
8.3
DECREASING PEAK RETENTION TIMES ............................................................................................ 23
8.4
DECREASING PEAK EFFICIENCY AND RESOLUTION......................................................................... 23
8.5
POOR PEAK EFFICIENCY AND RESOLUTION ..................................................................................... 24
8.6
UNIDENTIFIED PEAKS APPEAR......................................................................................................... 24
8.7
DECREASED DETECTION SENSITIVITY ............................................................................................. 24
8.8
COLUMN PROBLEMS ........................................................................................................................ 24
8.9
PEAK EFFICIENCY AND RESOLUTION ARE DECREASING ................................................................. 24
8.10
SYSTEM PROBLEMS.......................................................................................................................... 24
8.10.1
High Detection Background Caused by the System .............................................................. 24
8.10.2
No Peaks, Poor Peak Area Reproducibility or Unexpectedly Small Peak Area.................... 25
8.10.3
Incorrect or Variable Retention Times .................................................................................. 25
8.11
COLUMN CLEANUP........................................................................................................................... 25
8.11.1
High Salt Wash to Remove Ionic Components ..................................................................... 25
8.11.2
Organic Solvent Wash to Remove Non-Ionic Components .................................................. 25

DNAPac PA200 Document No. 065036 Page 4 of 25
SECTION 1 - INTRODUCTION
1.1 DNAPac PA200
The DNAPac PA200 is a pellicular anion exchange column designed specifically to provide high-resolution
separations of single stranded nucleic acids. The DNAPac PA200 provides n, n-1 resolution over a wide range of
oligomer lengths and can perform separations under a variety of denaturing conditions:
•High temperature, pH 8 or below
•High pH (12.4) at 30° or below
Because of the unique pH stability of the packing material, elevated pH conditions can be used to optimize
selectivity for specific oligonucleotides.
The packing material inside the DNAPac PA200 is composed of 130 nm quaternary amine functionalized
MicroBeads™ bound to an 8 µm solvent compatible, non-porous substrate. The non-porous substrate design
provides rapid mass transport resulting in narrow high efficiency peaks. The low column capacity, typical of non-
porous packings, is avoided by agglomerating functionalized MicroBeads to the surface of the substrate particle,
resulting in higher loading capacity than is possible with conventional non-porous materials, and good durability.
This produces a column with oligonucleotide resolution superior to columns using 2 to 3 µm resins.
Resin Characteristics:
Particle Size: 8 µm
Pore Size: non porous
Cross-linking: 55%
Ion exchange capacity: ~40 µeq/column
Latex Characteristics:
Functional Group: quaternary ammonium ion
Latex Diameter: ~130 nm
Latex Cross-link: 5 %
Typical Operating Parameters:
pH range: 4-10 unrestricted eluents
2.5-4 and 10-12.5, (Operation at these pH values require co-ion concentration to be
at least equimolar with hydroxide at high pH or H
+
at low pH)
Temperature: ≤85°C
Pressure: 3,000 psi
Organic Solvent Limit: 100% acetonitrile or methanol for cleaning
Typical eluents: High purity water (18.2 megohm-cm), sodium chloride, sodium perchlorate, buffers,
sodium acetate and sodium hydroxide
1.2 BioLC System (without Columns)
Table 1: System Components Recommended for DNA Analysis
Basic Gradient System Standard Gradient System
BioLC gradient pump, with degas BioLC gradient pump (degas recommended)
Chromatography oven with injection valve
and regulator assembly Autosampler
Column Oven
Absorbance detector (D
2
lamp for UV) Absorbance detector (D
2
lamp for UV)
EO1 eluent organizers EO1 Eluent organizers

DNAPac PA200 Document No. 065036 Page 5 of 25
1.3 Guard Column Use
A guard column is usually placed before the analytical column to prevent contaminants in the sample from eluting
onto the analytical column. The addition of the guard column increases the net column capacity, which translates
into an increase of about 20% in the retention times for isocratic runs. If a guard is added to a system running a
gradient method that was initially developed for an analytical column alone, the analytes will elute slightly later and
usually with slightly better resolution.
1.4 DNAPAC PA200 Anion Exchange Columns
Part Number Product Description
063000 DNAPac PA200, Analytical (4 x 250mm)
062998 DNAPac PA200, Guard (4 x 50mm)
1.5 DNAPac Column Family
There are two varieties of columns in the DNAPac column family. Both columns are non-porous anion exchangers
that provide high-resolution oligonucleotide separations. The choice of column depends upon the goal of the
separation. The DNAPac PA100 consists of a 13 µm substrate particle with 100 nm functionalized MicroBeads.
This column is available in a variety of formats and should be used when higher capacity is required and if scale-up
to semi-preparative scale separations is anticipated.
The DNAPac PA200 consists of an 8 µm substrate particle with 130 nm functionalized MicroBeads. This column
provides higher resolution than the DNAPac PA100. The DNAPac PA200 is operated at a lower flow rate than the
DNAPac PA100, thus less eluent is consumed during a run. In addition, the DNAPac PA200 has been
manufactured to provide greater stability to high pH at elevated temperature, although this combination is not
recommended.
Assistance is available for any problem that may be encountered during the shipment or operation of
DIONEX instrumentation and columns through the DIONEX North America Technical Call Center at 1-
800-DIONEX-0 (1-800-346-6390) or through any of the DIONEX offices listed in “DIONEX Worldwide
Offices.”
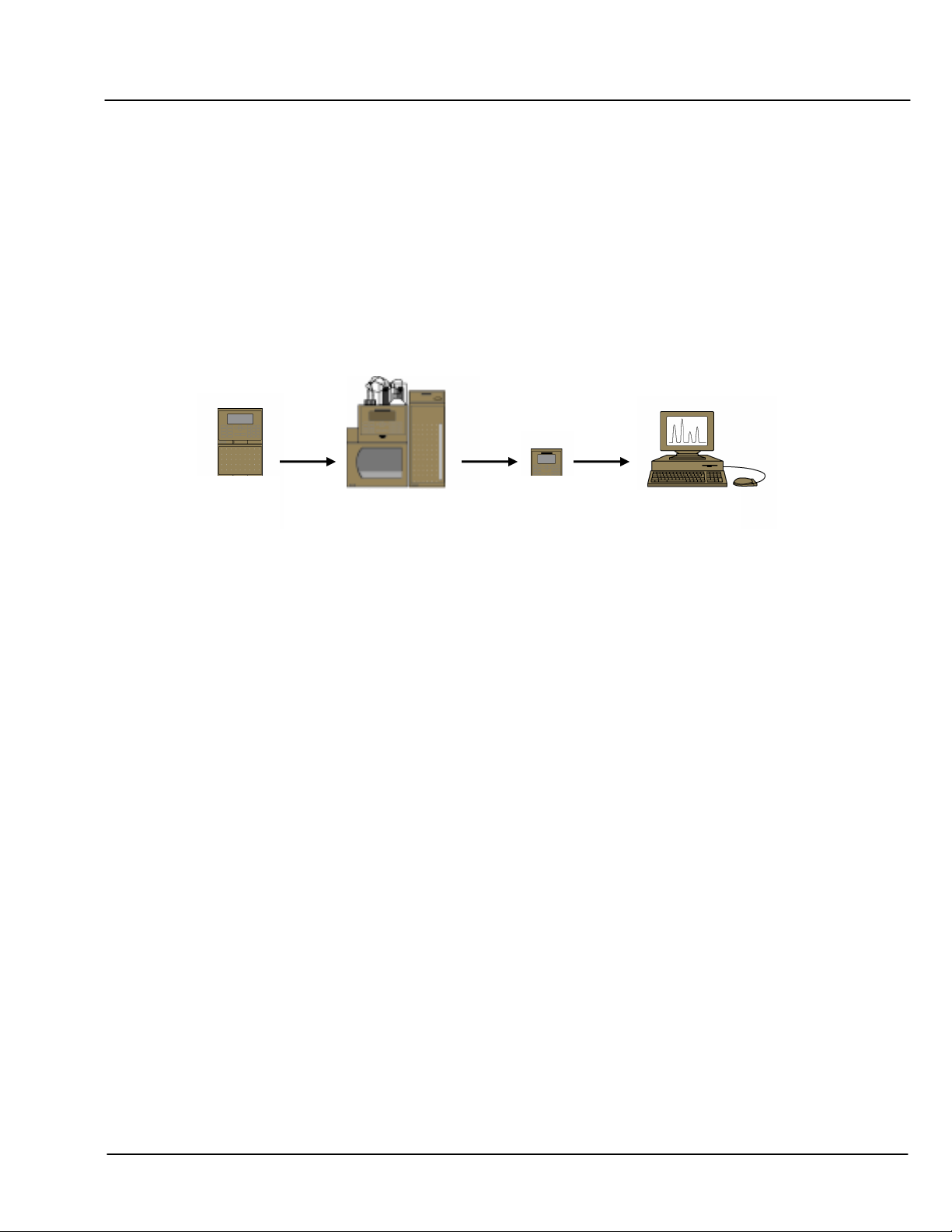
DNAPac PA200 Document No. 065036 Page 6 of 25
SECTION 2 - OPERATION AND SYSTEM REQUIREMENTS
2.1 System Requirements
Oligonucleotide separations with the DNAPac PA200 columns are optimized for use with NON-METALLIC
systems, such as the Dionex BioLC. The key issue is that the eluent flow path from reservoir to detector is metal-
free, because the salts used for oligonucleotide elution attack the metallic components of metallic pumps and
tubing. The released metals will irreversibly foul the column.
Each of the possible configurations offers multiple sampling options; however, consistently reproducible
quantification and an absence of disturbing artifacts are achieved best using an autosampler and “full loop”
injection mode. Reproducibility of retention time results can be enhanced by regulating the temperature of the
column using a column oven or thermal compartment.
Figure 1 Oligonucleotide System Configuration
2.2 System Operation Requirements
The oligonucleotide analysis systems should be configured with Dionex modules to provide the following
attributes:
a) All components of the fluid path are non-metallic, to eliminate column poisoning.
b) Mobile phase components are kept under helium or nitrogen to minimize out-gassing (bubble formation)
in the detector cell. On-line degassing of eluents may be provided with the eluent degas option on Dionex
pump modules.
c) Accurate reproducible flow and gradient generation at settings between 0.20 and 2.0 mL/min.
d) Minimal contribution to the background signal by contaminants from the system and reagents.
e) Thermostated column compartment for consistent temperature control of the guard and separation
columns.
f) Minimal system volumes (employ low volume unions and minimal tubing length).
•4-mm operation, liquid line inside diameter (I.D.) should be between .007” and 0.01”.
•2mm operation, liquid line inside diameter (I.D.) should be between .003” and 0.005”.
In both operations, PEEK tubing is preferred as it does not contribute to metal leaching.
Absorbance
Detector
Gradient
Pump Autosampler Thermal
Compartment
Data System

DNAPac PA200 Document No. 065036 Page 7 of 25
2.3 DNAPAC PA200 Column Operational Parameters
TABLE 2 Column Operational Parameters
pH = 4-10 (unrestricted eluents)
pH Range:
pH = 2.5- 4, and 10-12.5: Operation at these pH values require co-ion
concentration (e.g., Cl
-
or ClO
4
-
at high pH and Na
+
or NH
4
+
at low
pH) to be at least equimolar with hydroxide at high pH or H
+
at
low pH.
Temperature Limit: <85°C
Pressure Limit: 4,000 psi
Organic Solvent Limit: 100% Acetonitrile, or methanol, if required for cleaning.
Chaotrope Limit: 30% formamide, 6 M Urea.
Note: Use of these chaotropes will increase back pressure, and
reduce column lifetime.
Typical Eluents: High purity water (18 megohm-cm), sodium chloride, sodium
perchlorate, buffers, sodium acetate and sodium hydroxide.
Detergent
Compatibility: Nonionic, cationic or zwitterionic detergents.
CAUTION: Do not use anionic detergents. Anionic detergents will bind irreversibly to the column.

DNAPac PA200 Document No. 065036 Page 8 of 25
SECTION 3 - PURITY REQUIREMENTS FOR CHEMICALS
Reliable and reproducible results require eluents that are prepared consistently and are free from impurities.
3.1 Deionized Water
The de-ionized (DI) water, used to prepare eluents, should be Type I reagent grade water with a specific resistance
of 18 megohm-cm. The water should be free from ionized impurities, organics, microorganisms, and particulate
matter. Ultra Violet (UV) treatment in the water purification unit is recommended. Follow the manufacturer’s
instructions regarding the replacement of ion exchange and adsorbent cartridges. All filters used for water
purification must be free from UV absorbing components. Contaminated water in eluents causes high background
signals, gradient artifacts, and even sample degradation due to nucleases arising from microbial contamination.
3.2 Inorganic Chemicals
Inorganic chemicals of reagent grade or better should be used to prepare ionic eluents. Whenever possible,
inorganic chemicals that meet or surpass the latest American Chemical Society standard for purity should be used.
These products will include detailed lot analyses on their labels.
3.3 Solvents
Solvents can be added to the ionic eluents used in DNAPac PA200 columns to modify the ion exchange process.
The solvents used must be free from ionic impurities; however, since most manufacturers of solvents do not test for
ionic impurities, it is important that the highest grade of solvents available be used. Currently, several
manufacturers are making “ultra high” purity solvents that are compatible with HPLC and spectrophotometric
applications. These “ultra high” purity solvents will usually be of sufficient purity to ensure that your
chromatography is not affected by ionic impurities in the solvent. At Dionex, we have obtained consistent results
using High Purity Solvents manufactured by Burdick and Jackson or Optima Solvents by Fischer Scientific.
When using an ionic eluent with solvent, column generated back pressure will depend on the solvent used, the
concentration of the solvent, the ionic strength of the eluent, and the flow rate applied. The column backpressure
will also vary if the composition of the water-solvent mixture varies. The practical backpressure limit for the
DNAPac PA200 is 4,000 psi (27.6 MPa). The DNAPac PA200 can withstand common HPLC solvents in a
concentration range of 0-100%. Solvents and water should be premixed in concentrations which allow proper
mixing by the gradient pump and to minimize out-gassing. Ensure that all of the inorganic chemicals are soluble in
the highest solvent concentration to be used during the analysis.
Solvent-Water mixtures are usually specified with a volume to volume basis. If a procedure requires an eluent of
90% acetonitrile; prepare the eluent by adding 900 mL of acetonitrile to an eluent reservoir. Then add 100 mL of
deionized water, or eluent concentrate, to the acetonitrile in the reservoir. Using this procedure to mix solvents with
water will ensure that a consistent true volume/volume eluent is obtained. Premixing water with solvent will also
minimize the possibility of out gassing which causes bubble formation in the detector cell. If you choose to mix
eluents containing solvents with those that do not – the eluent degas option for the pump is highly recommended.
As a second choice, pre-degassing the eluents and covering the eluent reservoir with Helium gas to limit gas
dissolution into the eluents will help limit out-gassing.

DNAPac PA200 Document No. 065036 Page 9 of 25
SECTION 4 - QUALITYASSURANCE
The chromatograms in this section were obtained using a calibrated system that meets the operational parameters listed
in Section 2. Different systems will differ slightly in performance due to slight variations in column sets, system void
volumes, liquid sweep-out times, different component volumes, and laboratory temperature.
4.1 Certificate of Performance – Resin Batch Testing
Each batch of resin used for packing the DNAPac PA200 columns is tested to ensure reliable performance and
resolution. Separations of dT
19-24
with DNAPac PA200 columns, packed with both production and test resins, are
compared. This procedure ensures that resins with the highest quality are used, and produces consistent column
performance.
Eluent 1: 25 mM Tris pH 8
Eluent 2: 25 mM Tris pH 8, 1.25 M NaCl
Flow rate: 1.20 mL/minute
Detection: Absorbance (260 nm)
Injection: 25µL
Storage Solution: Eluent 2
Gradient: Time %1 %2 Comments
0.0 68 32 Equilibration solution
0.1 68 32 Equilibration solution
10.1 53.6 46.4 Gradient ramp 400 – 580 mM NaCl
10.11 35 60 Column wash start
11.11 35 60 Column Wash end
11.2 68 32 Re-Equilibration solution
14.12 68 32 End equilibration
Sample: dT
19-24
1.5 µg each / mL
CHART 1 Certificate of Performance
0.0 4.0 8.0 12.0
0
25
mA
260
Time (min)
dT19
dT20
dT21
dT22
dT23
dT24
Flow: 1.20 ml/min
46.4
60.0
46.4
60.0
1.25M NaCl:
32..0 %

DNAPac PA200 Document No. 065036 Page 10 of 25
4.2 Production Test Chromatograms
To guarantee that all DNAPac PA200 analytical columns meet high quality and reproducible performance
specification standards, all columns undergo the following production control test. Because gradient separation is
not an accurate test for determining column capacity and packing quality, an isocratic separation of seven inorganic
anions is employed to measure individual column performance utilizing a sodium carbonate, bicarbonate eluent.
The retention time of sulfate is used to measure the capacity of the column. Peak efficiency and peak symmetry of
sulfate are used to measure the packing quality of the column. Retention times and resolution of chloride, nitrate,
and phosphate are used to measure the overall selectivity of the column.
Eluent 1: 1.28mM NaHCO
3
1.35 mM Na
2
CO
3
Flow rate: 1.00 mL/minute
Detection: Conductivity (Suppressed)
Sample: 7 Anion Standard with (1/10 dilution)
Injection Volume: 25 µL
Storage Solution: 25 mM Tris pH 8, 1.25M NaCl
CHART 2 Production Test Chromatogram
0.0 2.0 4.0 6.0 8.0 10.0
0
30
Col # 102, 25.0
µS
Time (min)
F
-
Cl
NO
2-
Br
-
NO
3-
PO
4=
SO
4=
0.0 2.0 4.0 6.0 8.0 10.0
0
30
Col # 102, 25.0
µS
Time (min)
F
-
Cl
NO
2-
Br
-
NO
3-
PO
4=
SO
4=

DNAPac PA200 Document No. 065036 Page 11 of 25
SECTION 5 - METHODS DEVELOPMENT
5.1 Sample Cleanup
This table lists some sample preparation and matrix removal guidelines, for oligonucleotide samples, prior to
injection onto the DNAPac PA200 column.
TABLE 3 Sample Preparation and Matrix Removal Guidelines
Matrix
Interferent Effect Possible Removal
Halides High concentrations of salts in
the sample will affect the
retention time of analytes
Dialysis, dilution, ethanol precipitation,
cleanup with Reversed-phase
cartridges.
Anionic
Detergents
Will bind irreversibly to the
column
Dialysis, dilution, solid phase extraction
using the OnGuard RP Cartridge
5.2 Elution Order
The native elution order of oligonucleotide bases from the DNAPac PA200 phase using linear gradients
of NaCl or NaClO4 is as follows:
DNA > RNA (DNA is more retained than RNA)
Homopolymer Series:
pH 8: G > C > T > A
pH 12: G > T > C > A
NOTE: Poly-G will form extensive tetrad ladders at pH values below ~10.5. These are not readily disrupted,
even at 85°C in salt solutions.
Heteropolymer Series:
Elution is influenced by the base composition (especially % G +T), terminal base sequence, pH, solvent
concentration, and eluent salt. At pH 12 each T or G base contributes a negative charge from tautomeric
oxygen atoms; as pH shifts from 10.5 – 12.5, hydrogen bond interactions decrease yielding the expected
chromatographic patterns.
5.3 Effect of Salt Type on Oligonucleotide Elution
5.3.1 Eluent Strength
Sodium perchlorate (NaClO
4
) and sodium chloride (NaCl) are the two eluent salts used most commonly with
DNAPac columns. Sodium perchlorate is a stronger eluent than sodium chloride, so a higher concentration of
sodium chloride than of sodium perchlorate is required for any given separation. For example, typically ~0.2M
NaClO
4
will elute a 75-base oligonucleotide at pH 8, while ~0.7M NaCl would be required.
5.3.2 Loading Capacity
Column loading capacity is the maximum amount of a given oligonucleotide that can be loaded onto the
column before the peak shape starts to deteriorate. Column loading capacity is affected by the salt type. The
stronger the salt, the lower the loading capacity. Thus, use of NaClO
4
would result in peak broadening at a
lower sample loading concentration than a NaCl eluent.
DNAPac PA200 Document No. 065036 Page 12 of 25
5.4 Gradient slope
Phosphodiester oligonucleotides generally exhibit good peak shape when the gradient slope is ~15 mM/mL (NaCl)
or ~5 mM/mL (NaClO
4
). Higher values will generally result in shorter run times, but result in poorer resolution.
Conversely lower values may produce improved resolution, but also require longer run times.
5.5 Effect of pH and Solvent on Oligonucleotide Chromatography
Use of elevated pH offers two advantages over chromatography at neutral pH. First, elevated pH allows control of
hydrogen bonding interactions. At pH 11 and above, (pH 12.4 is the recommended upper limit for the DNAPac
columns), Watson-Crick and poly-G hydrogen bonds break. Hence, at high pH chromatographic analysis of
oligonucleotides with self-complementary sequences results in sharp, well-resolved peaks. Second, for each
Thymine (T) and Guanine (G) residue, an increase in oligonucleotide charge is generated with rising pH values due
to ionization of the tautomeric oxygen on these bases. Between pH 9 and 11, oxyanion formation on these bases
increases retention of oligonucleotides in proportion to the number of T and G residues on the molecule. This offers
opportunity to control of oligonucleotide selectivity with eluent pH.
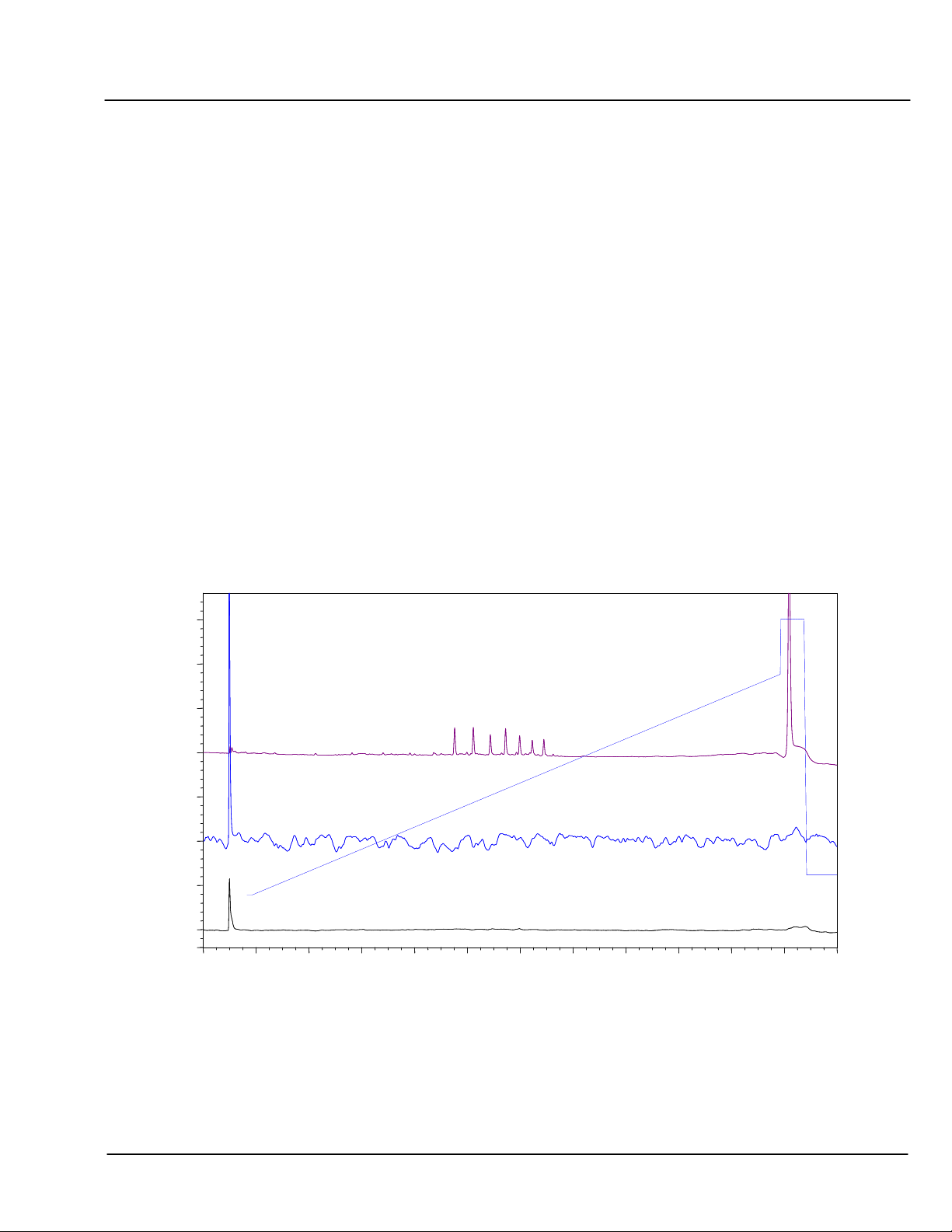
5.5.1 Effect of pH on Hydrogen Bond Interactions
In the chart below, the chromatography of a PdG
12-18
sample at 25°C and pH 8 (bottom trace) reveals the
absence of identifiable peaks. Under these conditions, poly-G tracts form tetrad ladders with 8 hydrogen bonds
between each set of bases. Chromatography at 85°C (pH 8, middle trace) dramatically increases the detector
noise compared to pH8 and 25°C, but still no identifiable peaks are eluted. However, at 25°C and pH 12 each
of the sample components are clearly eluted, and completely resolved from all the other components.
CHART 3 Controlling H-bond interactions in poly-G Tracts:
Comparison of High pH and High Temperature
0 8 16 24
0.0
40.0
d(G)
12-18
25 °C, pH8
d(G)
12-18
A 85 °C, pH8
d(G)
12-18
25 °C, pH12
mA
260
Time (min)
3
2
1
Flow: 1.20 ml/min
0.33M NaClO
4
:
33.4 %
66.7
75.0
36.4
0 8 16 24
0.0
40.0
d(G)
12-18
25 °C, pH8
d(G)
12-18
A 85 °C, pH8
d(G)
12-18
25 °C, pH12
mA
260
Time (min)
3
2
1
Flow: 1.20 ml/min
0.33M NaClO
4
:
33.4 %
66.7
75.0
36.4

DNAPac PA200 Document No. 065036 Page 13 of 25
5.5.2 Effect of pH on Retention
The next chart illustrates the influence of pH on oligonucleotide retention.
An oligonucleotide with base composition of G
6
C
5
A
5
T
9
was eluted with a
gradient of NaCl over 30 minutes at pH 6.5 to 12. Between pH 9 and 11, a
substantial increase in retention is observed. As shown in Figure 2, this is
due to the formation of an oxyanion on the tautomeric oxygen on each
G and T.
FIGURE 2
CHART 4 Effect of pH on Retention
5.5.3 Effect of Solvent on Retention
As shown in the next chart, addition of acetonitrile to the eluent will mask some of the native selectivity of the
DNAPac PA200, and reduce retention of oligonucleotides. In some cases, resolution of closely spaced or co-
eluting oligonucleotides may be assisted by adding solvent. These effects can be seen clearly by comparing
Chart 4 with Chart 5. The scales have been aligned to make this comparison easier.
CHART 5 Effect of Solvent on Retention
010 20
pH6.5
pH8
pH9
pH11
pH12
Time (min)
13.80’
14.35’
pH10
9.93’
6.02’
5.42’
4.61’
Flow: 1.20 ml/min
20% CH
3
CN
mA
260
010 20
pH6.5
pH8
pH9
pH11
pH12
Time (min)
13.80’
14.35’
pH10
9.93’
6.02’
5.42’
4.61’
Flow: 1.20 ml/min
20% CH
3
CN
mA
260
O
O
HN
N
O
R
N
N
R
HO
pH7 pH1
Time (min)
010 20 30
mA
260
22.74’
22.57’
15.04’
7.41’
6.88’
6.15’
Flow: 1.20 ml/min
1.25M
NaCl:
26.4 %
72.0
80.0
26.3
pH 6.5
pH 8
pH 9
pH 10
pH 11
pH 12
No CH3CN
Time (min)
010 20 30
mA
260
22.74’
22.57’
15.04’
7.41’
6.88’
6.15’
Flow: 1.20 ml/min
1.25M
NaCl:
26.4 %
72.0
80.0
26.3
pH 6.5
pH 8
pH 9
pH 10
pH 11
pH 12
No CH3CN
DNAPac PA200 Document No. 065036 Page 14 of 25
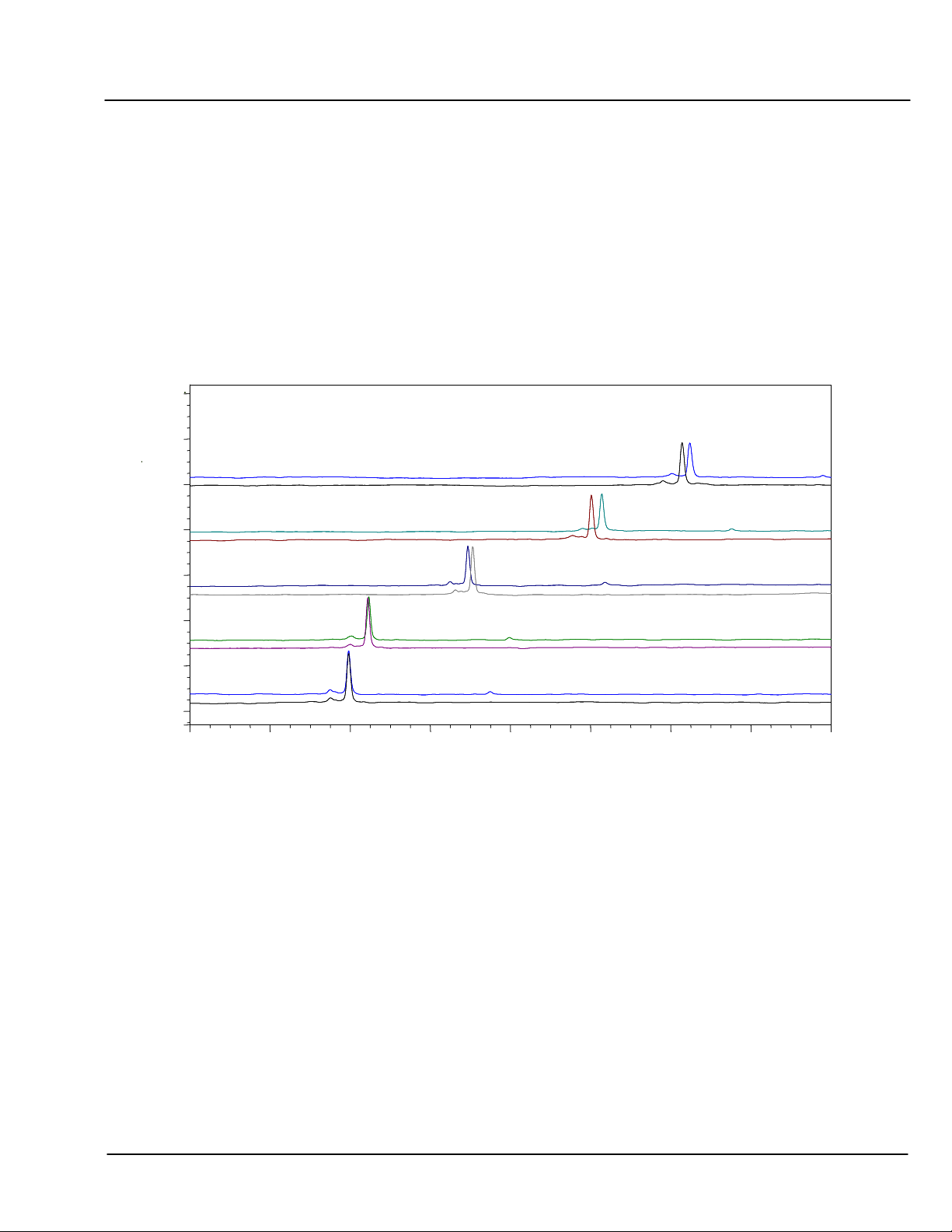
5.5.4 Effect of pH on Selectivity
The figure below illustrates the influence of pH on oligonucleotide selectivity. Here the elution patterns of two
23-base oligonucleotides differing only in their 5’ and 3’ terminal bases are compared between pH 9 and pH
11.
The top trace for each pair of chromatograms has an additional T at the 5’ end of the molecule, and lacks the 3’
A. The oligonucleotide chromatographed in the bottom trace lacks the 5’ T, but has the 3’A. At pH 9 - 9.5
(bottom 2 pairs of traces) these oligos are unresolved.
At pH 10, the 5’ TG-3’G oligonucleotide is eluted earlier than the 5’ G-3’ GA oligonucleotide, and the two are
only partially resolved. However, at pH 10.5 and 11 this elution order is reversed, due to the relative
contributions of T and A to retention at these pH values.
The base composition of these 23-base oligos is 5’ X-G
4
C
4
A
3
T
7
-Y 3’, and optimal resolution is observed at pH
10.5.
CHART 6 Effect of pH on Selectivity
75
412
0
mAU
Time (Min)
10
9
8
7
6
5
4
3
2
WVL:260 nm
1
Flow: 1.20 ml/min
X=GA, Y=TGA, pH 9
X=TGA, Y=TG, pH 9
X=GA, Y=TGA, pH 9.5
X=TGA, Y=TG, pH 9.5
X=GA, Y=TGA, pH 10
X=TGA, Y=TG, pH 10
X=GA, Y=TGA pH 10.5
X=TGA, Y=TG, pH 10.5
X=GA, Y=TGA, pH 11
X=TGA, Y=TG, pH 11
56 7 910118
75
412
0
mAU
Time (Min)
1010
99
88
77
66
55
44
33
22
WVL:260 nm
1
1
Flow: 1.20 ml/min
X=GA, Y=TGA, pH 9
X=TGA, Y=TG, pH 9
X=GA, Y=TGA, pH 9.5
X=TGA, Y=TG, pH 9.5
X=GA, Y=TGA, pH 10
X=TGA, Y=TG, pH 10
X=GA, Y=TGA pH 10.5
X=TGA, Y=TG, pH 10.5
X=GA, Y=TGA, pH 11
X=TGA, Y=TG, pH 11
56 7 910118
DNAPac PA200 Document No. 065036 Page 15 of 25
5.6 Effect of Temperature on Oligonucleotide Retention
Elevated temperature is often used to limit or eliminate Watson-Crick, and poly-G hydrogen bonding within, and
between oligonucleotides that have self-complementary sequences.
NOTE: Dionex does NOT recommend combining the use of elevated temperatures with high pH elution
systems. Such conditions will accelerate degradation of the DNAPac PA200 stationary phase.
At relatively low pH, 9 or below, increased temperature may have mixed effects on nucleic acid retention.
Nucleoside monophosphates and very short, 2-3 base, oligonucleotides may exhibit decreased retention times at
elevated temperatures. Nucleoside triphosphates and oligonucleotides greater than a few bases long usually exhibit
increased retention as the temperature increases. The chart below illustrates the influence of increased temperature
at constant pH (8). As the chromatographic temperature increases, retention of the oligonucleotides also increases,
in this case by an average of ~ 2.7 min per 10°C.
CHART 7 Effect of Temperature on Retention of Oligonucleotides
d(AC)
10-11
: pH 8, 5 mM NaClO
4
/ mL
0.0 4.0 8.0 12.0 16.0 20.0
0
100
d(AC)x-xi 25 °C
d(AC)x-xi 35 °C
d(AC)x-xi 45 °C
d(AC)x-xi 55 °C
mAU
min
4
3
2
1
WVL:260 nm
Flow: 1.20 ml/min
0.33M NaClO4:
21.2 %
51.5
75.0
22.1

DNAPac PA200 Document No. 065036 Page 16 of 25
5.7 Effect of Terminal Base on Selectivity
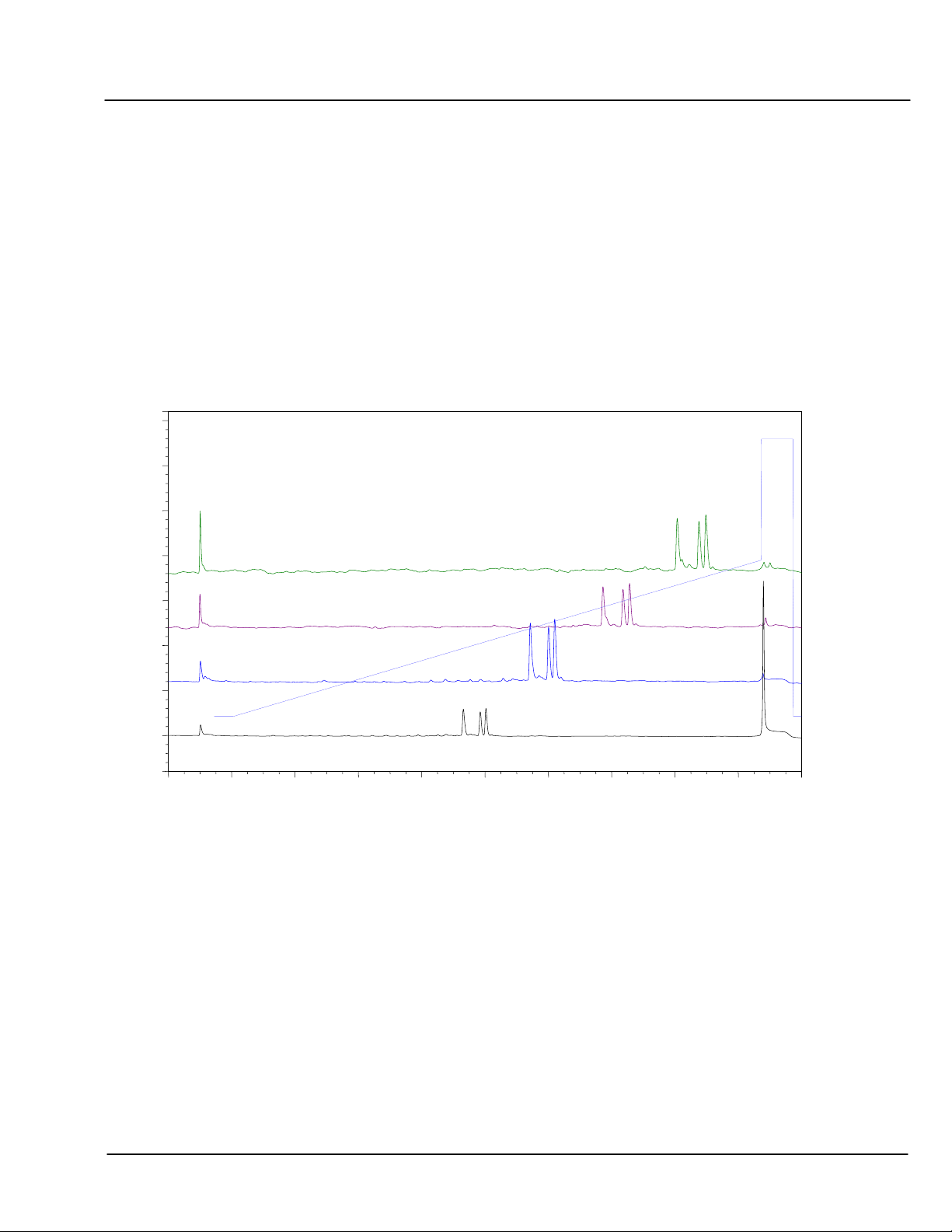
5.7.1 Selectivity in Sodium Chloride (NaCl) Gradients
The influence of the 5’ and 3’ terminal base on retention is shown below for NaCl eluents. These examples
consist of mixed-base oligonucleotide (ODN) 25 mers with identical sequence except for the 3’ and 5’ terminal
bases. The samples are chromatographed at pH 9, 10 & 11.
In each panel, the top 4 traces show elution of ODNs with identical 5’ termini, and altered only at the 3’ base.
The bottom 4 traces show elution of ODNs with identical 3’ termini, and altered only at the 5’ base. The
middle trace is common to both sets.
Using NaCl as the salt, all of the ODNs differing only at the 3’ base are resolved at pH 9 or 10. The ODNs
differing only at the 5’ base are also at least partially resolved at pH 9 or 10. In each case, C contributes the
least to retention at either the 5’ or 3’ end, and G contributes the most.
The relative contributions of A and T at either end are pH dependent. At pH 9, A contributes more than T but
at the higher pH values it contributes less.
Addition of solvent (e.g., CH
3
CN) tends to reduce retention, and minimize hydrophobic interactions. This may
in some cases improve selectivity and resolution, (chart not shown).
CHART 8 Effect of pH on Retention (by Terminal Base):
15 mM/mL NaCl gradient, 0% CH
3
CN, 5’X-G
6
C
3
A
5
T
9
-Y3’, pH 9-11
5.5 6.5 7.5 8.5
-1
140 mA
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 9 pH 11
21.5 22.5 23.5 24.5
-1
140 mA
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 10
13.5 14.5 15.5 16.5
-1
140 mA
Time (min)
7
6
5
4
3
2
1
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
Flow: 1.20 ml/min
5.5 6.5 7.5 8.5
-1
140 mA
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 9 pH 11
21.5 22.5 23.5 24.5
-1
140 mA
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 10
13.5 14.5 15.5 16.5
-1
140 mA
Time (min)
7
6
5
4
3
2
1
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
Flow: 1.20 ml/min

DNAPac PA200 Document No. 065036 Page 17 of 25
5.7.2 Selectivity in Sodium Perchlorate (NaClO
4
) Gradients
When NaCl eluent is replaced with NaClO
4
(see below), the retention differences are less pronounced, and the
effect of pH on retention is also reduced. However, all of the ODNs with 3’ base substitutions are again
resolved, and those with 5’ substitutions are at least partially resolved, at pH 9 or 10. Addition of solvent to
NaClO
4
eluent will reduce retention and minimize hydrophobic interactions, resulting in smaller selectivity
changes due to terminal base differences at these pH values.
CHART 9 Effect of pH on Retention (by Terminal Base):
5 mM/mL NaClO
4
gradient, 0% CH
3
CN, 5’X-G
6
C
3
A
5
T
9
-Y3’, pH 9-11
7 8 9 10
-1
99 mAU
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 9 pH 10
9.5 10.5 11.5 12.5
-1
99 mAU
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 11
13 14 15 16
-1
99 mAU
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
7 8 9 10
-1
99 mAU
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 9 pH 10
9.5 10.5 11.5 12.5
-1
99 mAU
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G
pH 11
13 14 15 16
-1
99 mAU
Time (min)
7
6
5
4
3
2
1
Flow: 1.20 ml/min
X=C, Y=A
X=A, Y=A
X=T, Y=A
X=G, Y=A
X=C, Y=T
X=C, Y=C
X=C, Y=G

DNAPac PA200 Document No. 065036 Page 18 of 25
5.8 Application-Specific Mobile Phase Recommendations
From the observations detailed in the preceding sections, the following suggestions can be made:
5.8.1 Eluent Systems Minimizing Base-Specific Retention
For synthetic ODNs where the goal is to evaluate purity, determine the coupling efficiency, or purify the full-
length component from “n-1” and “n+1” impurities in the sample, eluent systems minimizing base-specific
retention would produce the best results. Hence, solvent-containing NaClO
4
eluent at pH 9 or below, where
pH-induced ionization is further minimized, would be the logical choice.
5.8.2 Eluent Systems Maximizing Base-Specific Retention
When multiple possible ODNs of similar length in the same solution must be resolved, eluents maximizing
base-specific retention would provide the best probability of success. Examples of such samples include:
Identification of all primers in a multiplex PCR amplification cocktail, QA / QC of multiple primers in
amplification-based diagnostic kits, identification of the different components in “n-1” or “n+1” impurities
when troubleshooting nucleic acid synthesis protocols. For these applications, NaCl without solvent, at pH
values between 9 and 11 would be more likely to produce the desired separations.
5.8.3 Exploitation of Interactions Between The stationary phase and ODN Derivatives
When hydrophobic interactions between some bases and the phase are suspected, interactions between the
phase and additional ODN derivatives may also be exploited. Examples of such derivatives include numerous
fluorescent dyes, and the “Trityl” group used to protect the oligo from unwanted base additions at each
elongation step during ODN synthesis.

DNAPac PA200 Document No. 065036 Page 19 of 25
SECTION 6 - APPLICATIONS
6.1 Denaturing Conditions for Control of Secondary Structure
Single-stranded nucleic acids may contain inter-, and/or intra-, strand hydrogen bonding. Such interactions, if
sufficiently strong, result in spurious peaks and a general inability to distinguish between the oligonucleotide
components in the sample. There are three common methods to restrict these interactions; high temperature,
addition of chaotropic agents such as urea or formamide, and use of high pH. Both the temperature used and the
concentration of chaotropic agent used depend upon the extent of hydrogen bonding. For pH, values between pH
11-12.4 are effective at controlling both Watson-Crick, and non Watson-Crick oligonucleotide interactions.
While the DNAPac PA200 can be used with any of the above methods for controlling secondary structure, there are
certain considerations that should be taken into account when deciding which approach to use:
a) The use of a chaotropic agent, such as formamide or urea, tends to reduce the lifetime of the column.
b) The use of elevated temperature tends to reduce the lifetime of the column
c) Elevating the temperature of the DNAPac PA200 will increase the retention time of the oligonucleotide.
This means that more eluent will be required to elute the oligonucleotide, and thus the amount of salt
eluting with it will be increased
d) Increasing the pH of the eluent will also generally increase retention of oligonucleotides, but in a manner
that allows control of oligonucleotide selectivity.
6.2 Effect of High Temperature and High pH on Column Lifetime
The combination of both high temperature and high pH reduces the useful life of the DNAPac columns, as shown
in the Chart below. DNAPac PA100 is more susceptible to column degradation when the combination of high
temperature and high pH are employed. However, even the DNAPac PA200 shows some phase degradation when
operated at 65
0
C and pH 12, and this combination is not recommended.
CHART 10 Summary of Phase Stability in Alkali: DNAPac PA100 vs. PA200
0.0
0.2
0.4
0.6
0.8
1.0
0 20 40 60 80 100 120
Hours Exposure
Fraction of Initial Capacit
y
25 °C pH 12
45 °C pH 12
65 °C pH 12
Isocratic Column Degradation:
pH 12.4 vs. Temperature: DNAPac PA100
0.0
0.2
0.4
0.6
0.8
1.0
0 20 40 60 80 100 120
Hours Exposure
25 °C pH 12
45 °C pH 12
65 °C pH 12
Isocratic Column Degradation:
pH 12.4 vs. Temperature: Prototype
Fraction of Initial Capacity
0.0
0.2
0.4
0.6
0.8
1.0
0 20 40 60 80 100 120
Hours Exposure
Fraction of Initial Capacit
y
25 °C pH 12
45 °C pH 12
65 °C pH 12
Isocratic Column Degradation:
pH 12.4 vs. Temperature: DNAPac PA100
0.0
0.2
0.4
0.6
0.8
1.0
0 20 40 60 80 100 120
Hours Exposure
25 °C pH 12
45 °C pH 12
65 °C pH 12
Isocratic Column Degradation:
pH 12.4 vs. Temperature: Prototype
Fraction of Initial Capacity

DNAPac PA200 Document No. 065036 Page 20 of 25
6.3 Phosphodiester Analysis
6.3.1 Sodium Perchlorate Eluent Systems
The following separation represents a good starting guideline for developing sodium perchlorate (NaClO
4
)
based methods for longer oligonucleotides.
In this example, phosphorylated deoxycytosine oligomers, 19 – 24 bases long, were injected onto a DNAPac
PA200 column and eluted according to the conditions listed below. At pH 8, this gradient is effective for
resolving the “full-length” oligonucleotide phosphodiesters, up to 25 bases long, from the n-1 components.
The same gradient of 5 mM NaClO
4
per mL of eluent can also be used to resolve full length from n-1
components between pH 8 and pH 12, using other buffers, e.g., AMPS, Na
3
PO4, etc.
CHART 11 N, N-1 Separation of Phosphorylated Deoxycytosine Oligomers
Conditions: 22 minute gradient from 70 to 202 mM NaClO
4
in 20 mM Tris buffered eluent at pH 8.
Flow rate: 1.2 mL/minute.
Injection volume: 6 µL.
Sample: 1 A
260
/mL solution of phosphorylated deoxycytosine oligomers, Temperature: 25°C
0
4
80
12
16
20
24
0.
20.
P(dC)
19-24
mA
260
Time
(
min
)
This manual suits for next models
2
Table of contents
Other Dionex Analytical Instrument manuals



















