2.2 Updated January 2006
Contents
2.0 Introduction................................................................................................................3
2.1 Purging.......................................................................................................................3
2.2 Power-up....................................................................................................................4
2.3 Launching the software..............................................................................................6
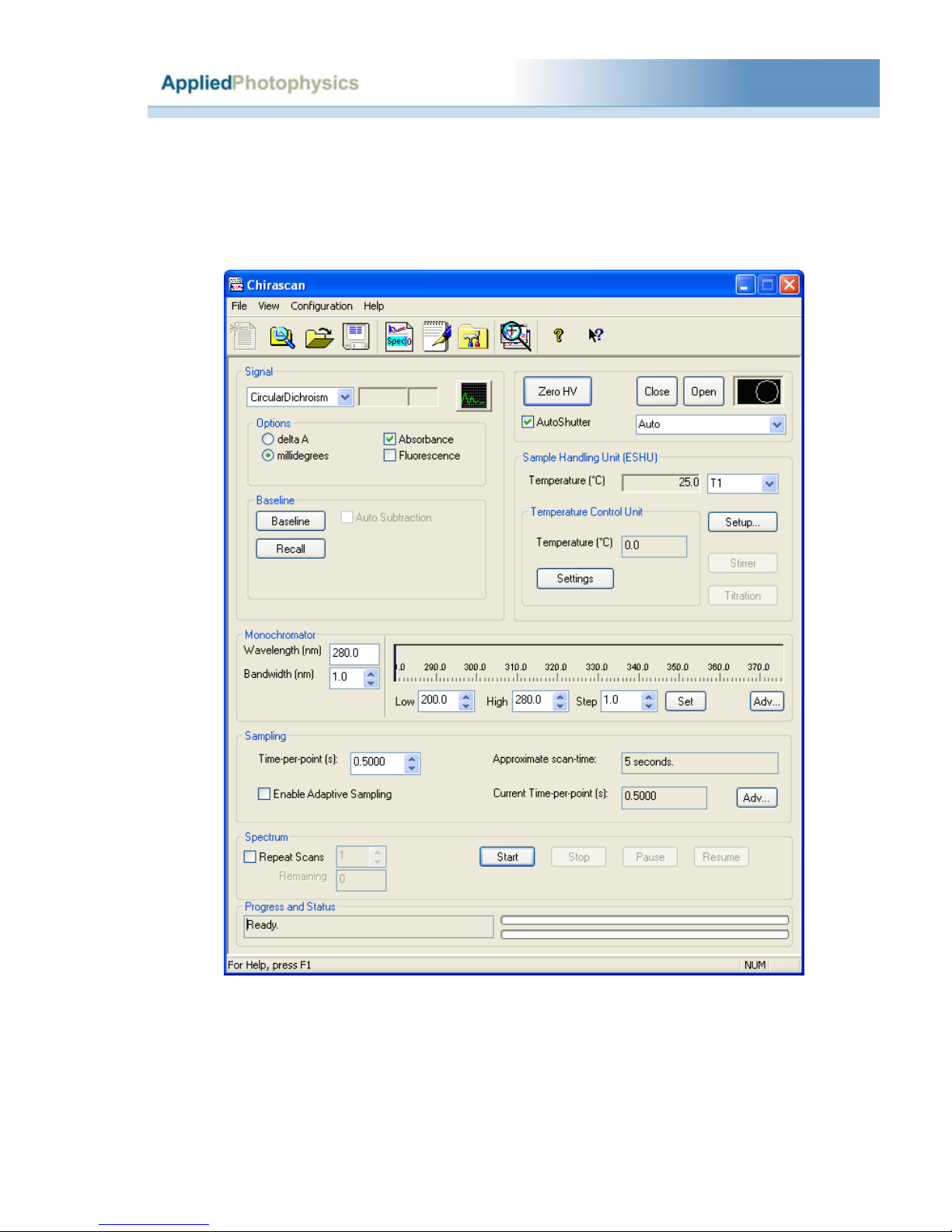
2.3.1 Measuring a CD baseline and spectrum..................................................................7
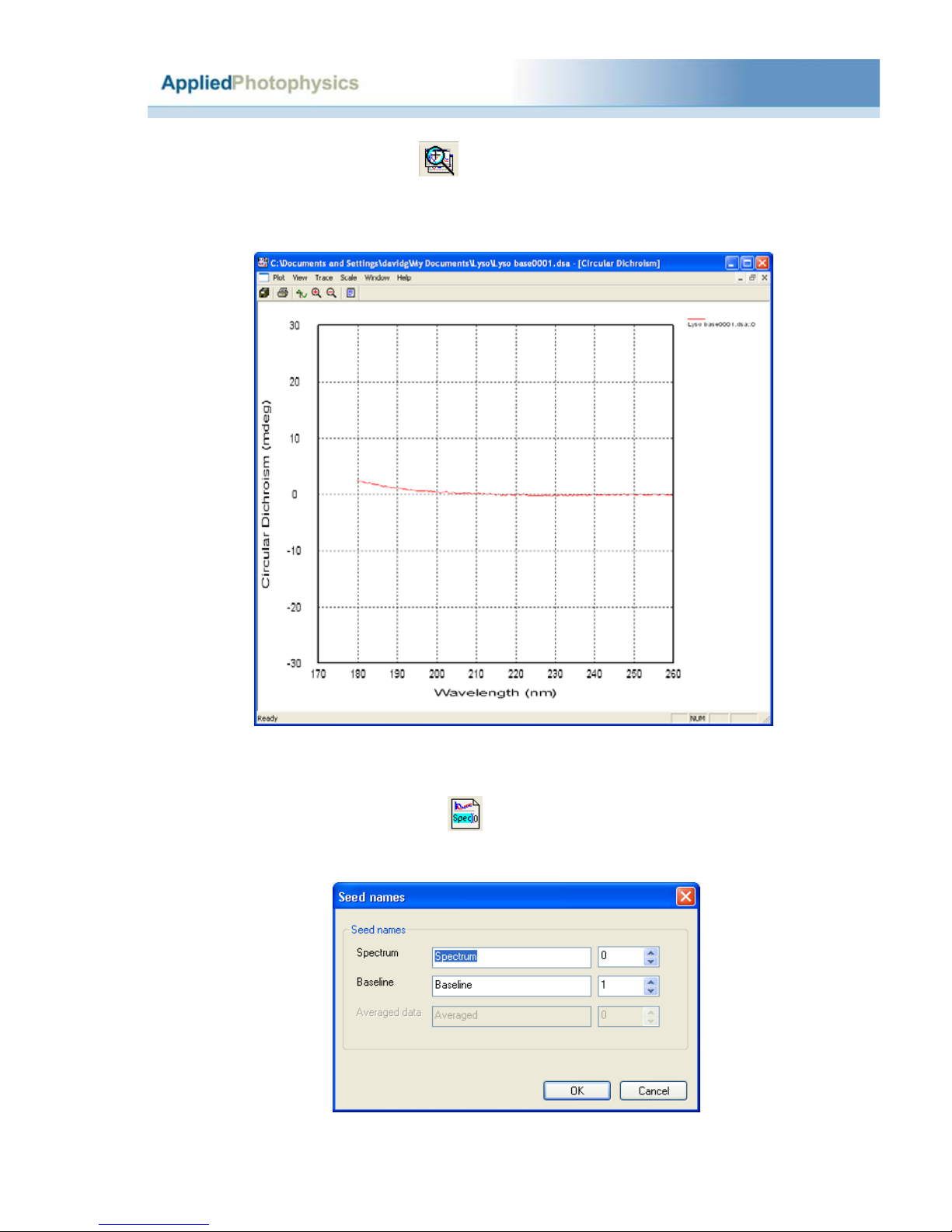
2.3.1.1 The CD baseline...................................................................................................7
2.3.1.2 The CD spectrum...............................................................................................10
2.4 Viewing and manipulating spectra and traces..........................................................11
2.4.1 Viewing spectra ....................................................................................................12
2.4.2 Comparing spectra................................................................................................13
2.4.3 Manipulating spectra.............................................................................................14
2.4.4 Example of spectrum manipulation ......................................................................21
2.5 Saving data...............................................................................................................26
2.5.1 Converting data for use with third-party programs...............................................27
2.6 Printing.....................................................................................................................28
2.7 Operating notes and hints.........................................................................................28
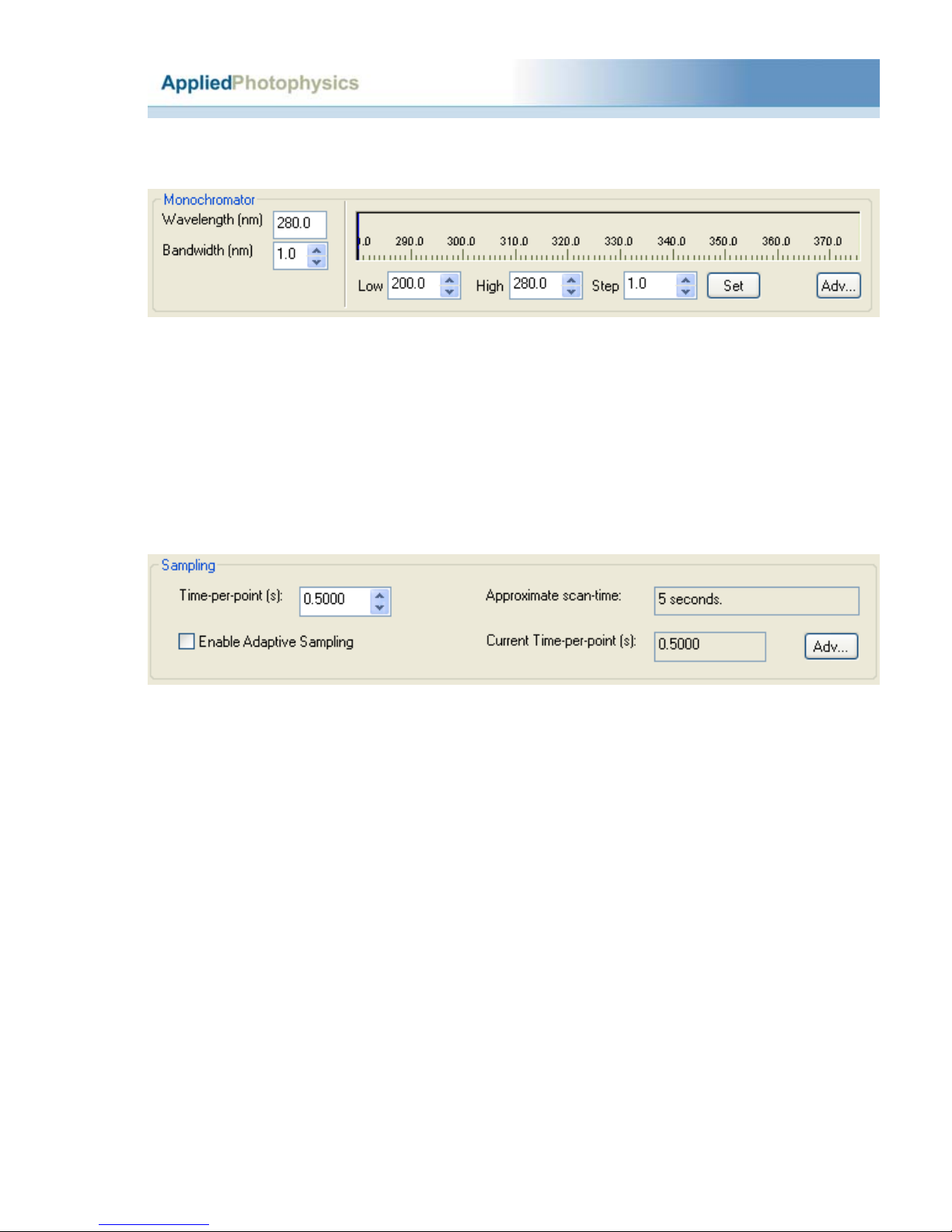
2.7.1 Selecting Step Size (SS)........................................................................................28
2.7.2 Selecting Spectral Bandwidth (SBW)...................................................................28
2.7.3 Selecting pathlength and concentration.................................................................29
2.7.4 Selecting time per point ........................................................................................29
2.7.5 Nitrogen purge flow-rate.......................................................................................30
2.7.6 Measuring protein spectra.....................................................................................30
2.8 Example spectra.......................................................................................................31
2.8.1 Alcohol dehydrogenase.........................................................................................32
2.8.2 Bovine serum albumin..........................................................................................33
2.8.3 Cytochrome C.......................................................................................................34
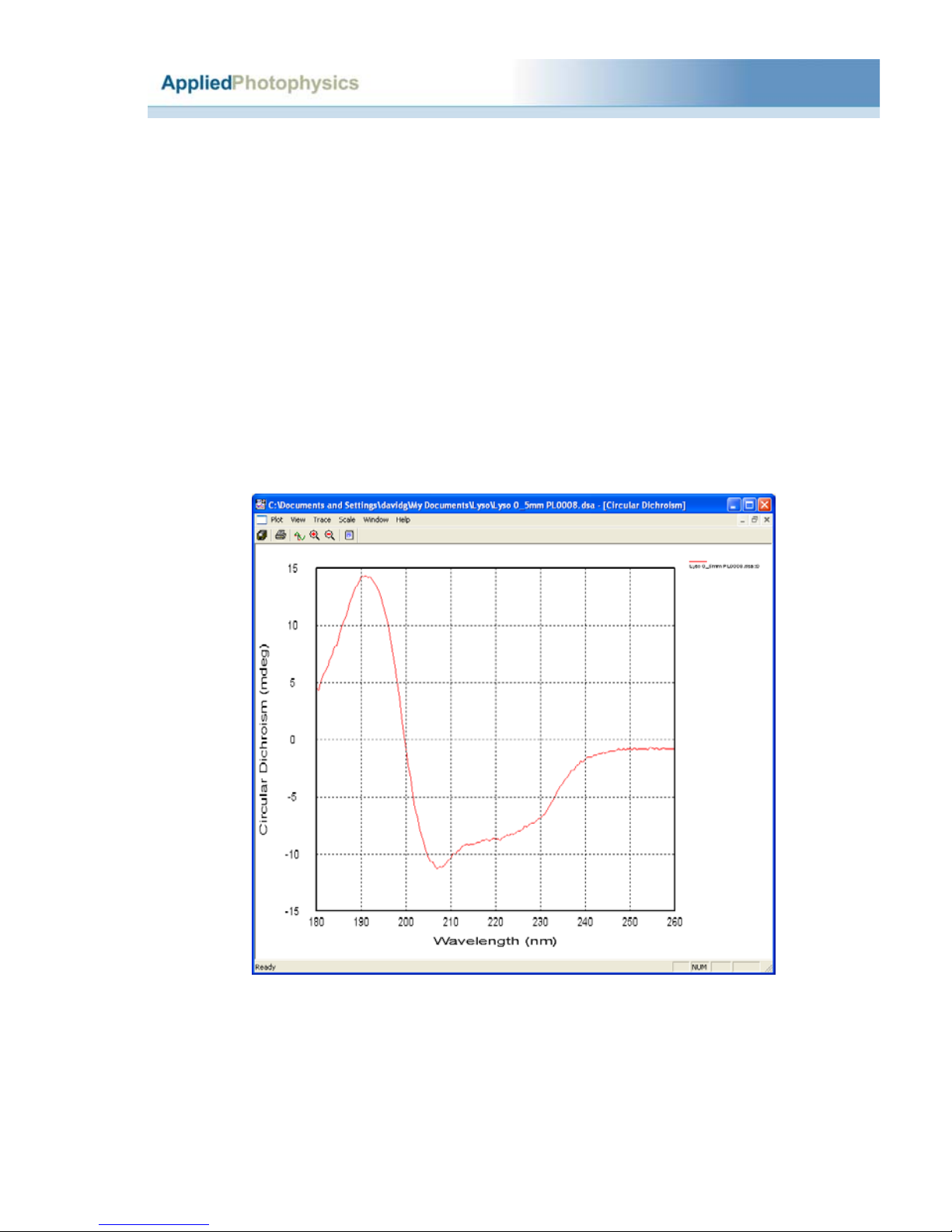
2.8.4 Lysozyme..............................................................................................................35
2.8.5 Vitamin B12..........................................................................................................36
2.8.6 Tris(ethylenediamine) cobalt chloride ..................................................................37
2.8.7 Camphor sulphonic acid........................................................................................38
2.8.8 (R)-3-methylcyclopentanone ................................................................................39
2.9 Troubleshooting.......................................................................................................40
2.10 Notes......................................................................................................................41