Clontech PT3139-1 User manual

Advantage®-HF PCR Kit
User Manual
(PT3139-1)
Catalog #K1909-1, -y
Storage conditions: –20°C
FOR RESEARCH USE ONLY
(PR76834)

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
2 Version # PR76834 FAX:415-424-1064 or 800-424-1350
Table of Contents
I. Introduction 3
II. List of Components 7
III. Additional Materials Required 8
IV. Advantage-HF PCR Kit Protocol 9
A. General Considerations 9
B. Control PCR Reactions 11
C. Recommended Cycling Parameters 12
D. Amplification of Longer Fragments with the Advantage Buffer 13
E. Recommendations for Electrophoresis 13
V. Troubleshooting Guide 14
VI. References 18
VII. Related Products 19
Notice to Purchaser
AlicenseunderU.S.patents4,683,202,4,683,195,and4,965,188ortheirforeigncounterparts,ownedbyHoffmann-
La Roche and F. Hoffmann-La Roche Ltd. (“Roche”), has an up-front fee component and a running-royalty
component. The purchase price of this product includes limited, non-transferable rights under the running-royalty
component to use only this amount of the product to practice the Polymerase Chain Reaction (“PCR”) and related
products described in said patents solely for the research and development activities of the purchaser when this
product is used in conjunction with a thermal cycler whose use is covered by the up-front fee component. Rights to
theup-frontfeecomponentmustbeobtainedbytheend-userinordertohaveacompletelicense.Theserightsunder
the up-front fee component may be purchased from Perkin-Elmer or obtained by purchasing an authorized thermal
cycler. No right to perform or offer commercial services of any kind using PCR, including without limitation reporting
the results of purchaser’s activity for a fee or other commercial consideration, is hereby granted by implication or
estoppel. Further information on purchasing licenses to practice the PCR process may be obtained by contacting
the Director of Licensing at the Perkin-Elmer Corporation, 850 Lincoln Centre Drive, Foster City, CA 94404 or at
Roche Molecular Systems, Inc., 1145 Atlantic Avenue, Alameda, CA 94501.
Thisproductissold underlicensingarrangementswith F.Hoffmann-LaRocheLtd., RocheMolecularSystems,Inc.,
and the Perkin-Elmer Corporation.
Advantage-HF cDNA Polymerase Mix is covered by U.S. Patent No. 5,436,149. Foreign patents pending.
TaqStart Antibodies are licensed under U.S. Patent No. 5,338,671 and corresponding patents in other countries.
CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 3
I. Introduction
The Advantage®-HF (High-Fidelity) PCR Kit is a KlenTaq-based system de-
signed to deliver
Pfu
-like fidelity in the amplification of cDNA or genomic
templates.
High fidelity
and
efficiency
While
Pfu
polymerase is known for its exceptional fidelity, it suffers from subop-
timalefficiencythatcanbeproblematicinmanyPCRapplications.Incontrast,the
Advantage-HF PCR Kit offers
Pfu
-like fidelity combined with the efficiency
required to amplify DNA fragments of up to 2.5 kb. These benefits are the result
of reformulation of several components in CLONTECH’s Advantage PCR
Enzyme Systems. The Advantage-HF Polymerase Mix combines KlenTaq (a 5'-
exonuclease-deficient variant of
Taq
polymerase) with a proofreading poly-
merase and TaqStartTM Antibody to provide a superior level of specificity.
Advantage-HF thus combines the benefits of
Pfu
and the Advantage Enzyme
System to deliver a high-fidelity enzyme system.
The HF Advantage
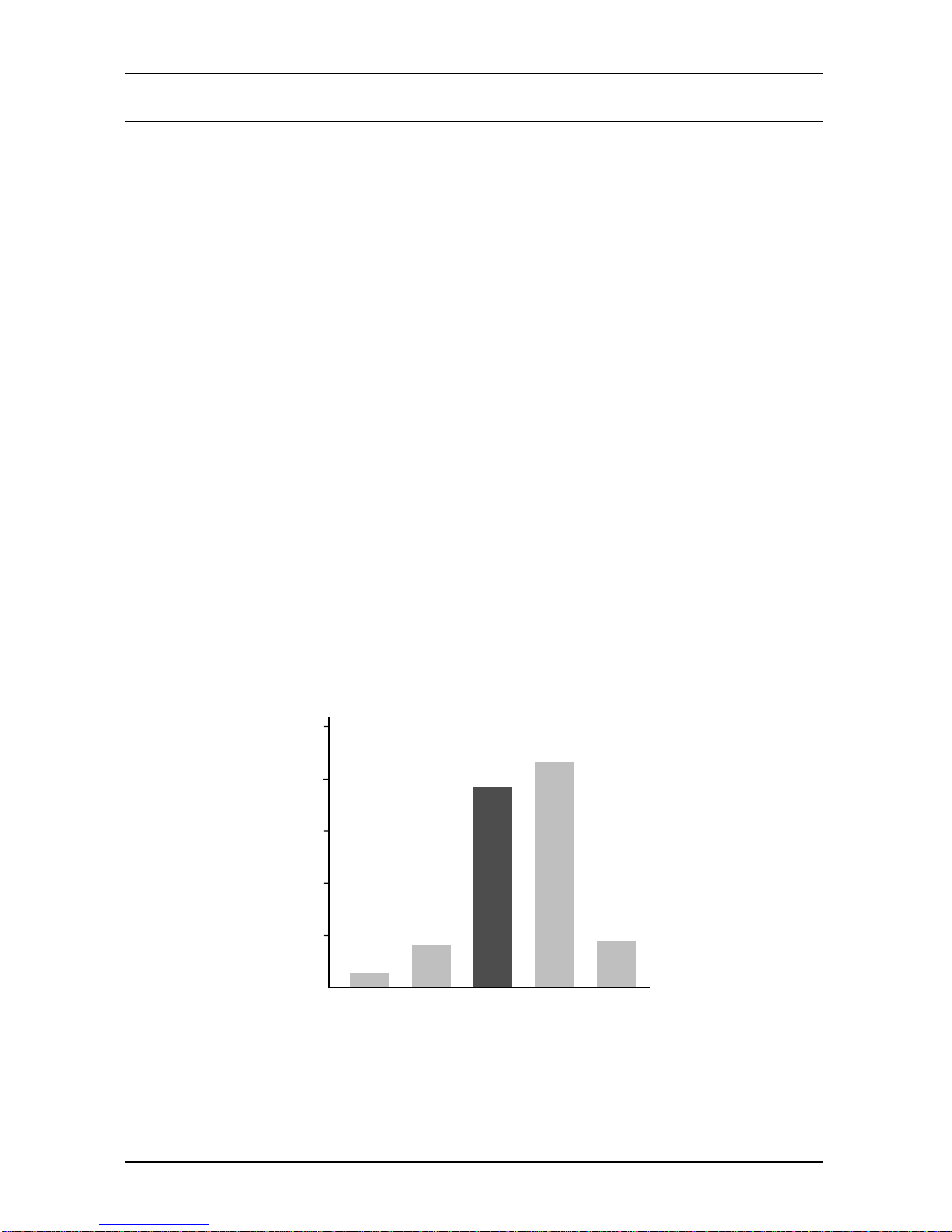
TheaccuracyofAdvantage-HFiscomparedtootherenzymesandenzymemixes
in Figure 1. Using a genetic assay that measures nucleotide misincorporation,
Advantage-HFrivals
Pfu
infidelity. Thisfidelityassayisbased onamplificationof
an
E. coli
ribosomal protein gene (Mo
et al.
, 1991). Mutations in this gene often
confer streptomycin resistance on the host. Upon introduction of the amplified
DNA into
E. coli
, the ratio of total transformants to streptomycin resistant
Figure 1. Comparison of fidelity of Advantage-HF and other PCR systems. The fidelity of
Advantage-HFcompares favorably with that of the
Pfu
enzyme and is significantly higher than that of
other enzyme systems. Accuracy assay is based on 25 cycles of amplification (see text).
0
100
200
300
400
500
19
67
385
435
71
Taq Advantage Advantage
HF Pfu
Taq
+
Pwo
Enzyme
Accuracy
(total transformants/strep
r
transformants)

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
4 Version # PR76834 FAX:415-424-1064 or 800-424-1350
I. Introduction
continued
transformants provides a comparative measure of PCR fidelity. The fidelity of
Advantage-HF was confirmed by sequencing (Table I). The high level of fidelity
delivered by the Advantage-HF system increases confidence in sequence de-
rived from PCR products and is beneficial in a variety of PCR applications,
including expression studies of amplified full-length cDNAs, generation of cDNA
libraries, RACE, and analysis of homologous genes amplified with degenerate
primers.
TABLE I. FIDELITY OF ADVANTAGE-HF BASED ON SEQUENCING DATA
Error ratea
Enzyme (per 100,000 bp)
Taq
180b
Advantage-HF 2.4
adetermined with individual clones after 25 PCR cycles
b agrees with published data (Ling
et al.
, 1991; Cariello
et al.
, 1991)
High-fidelity amplification of cDNA and Genomic templates
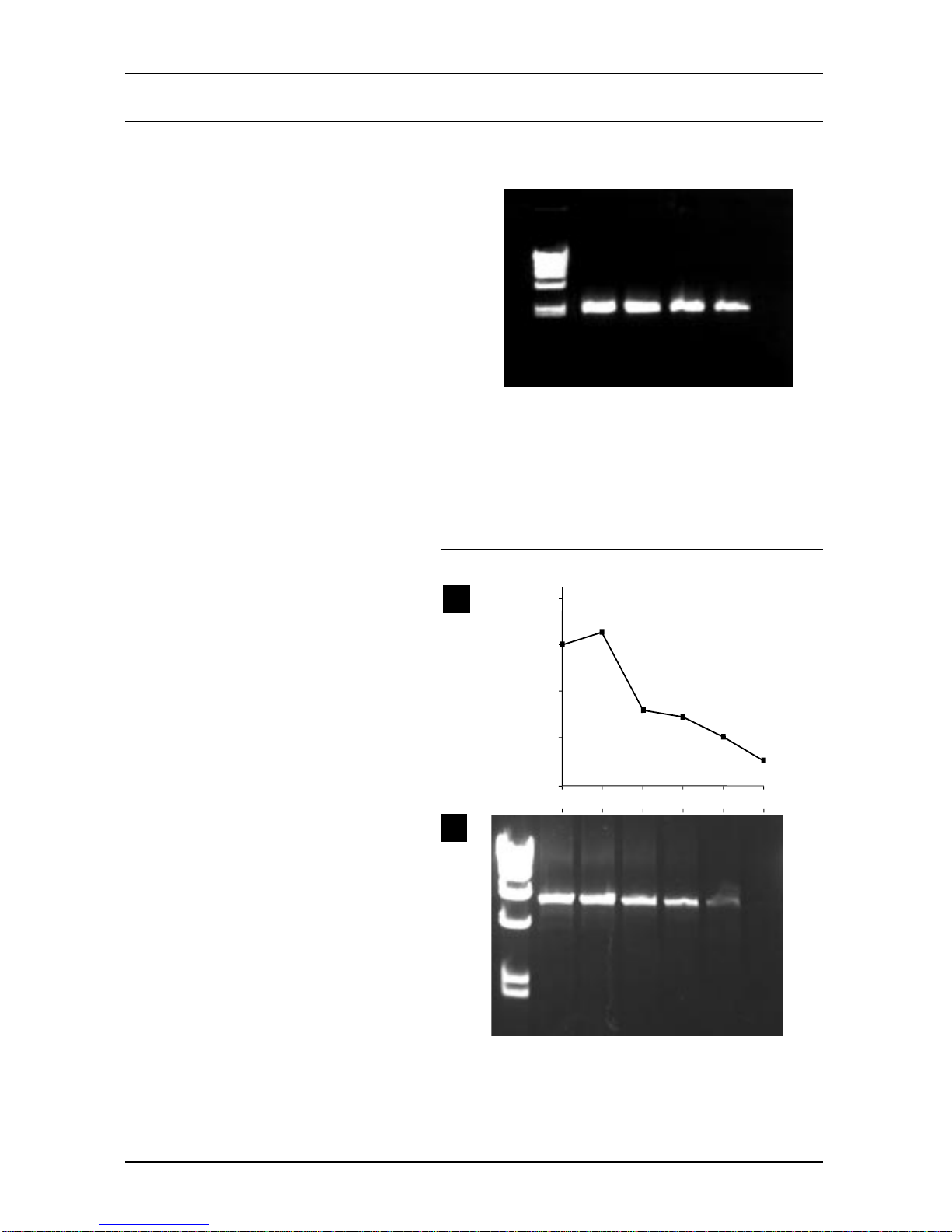
Advantage-HF was used to amplify several cDNA templates of different lengths
(Figure 2). Although amplification of the longest template yielded a reduced
amount of product (Lane 6), this amplified product contains a higher percentage
ofaccuratecopies—nearly6-foldhigherthanAdvantage,and20-foldhigherthan
Taq
, according to Figure 1.
Figure2.Advantage-HFamplificationofcDNAfragments.Severalfragmentswereamplifiedfrom
HumanPlacentacDNAunderstandard(Lanes1,3&5)andhigh-fidelityPCRconditions(Advantage-
HF; Lanes 2, 4 & 6). M: λ/
Hin
d III DNA size markers. Lanes 1 & 2: 0.5-kb fragment of glycerol 3-
phosphate dehydrogenase gene. Lanes 3 & 4: 1.3-kb fragment of transferrin receptor gene. Lanes 5
& 6: 2.5-kb fragment of lactoferrin gene. Cycling parameters: 30 sec at 94oC; 30 x (30 sec at 94oC,
5 min at 68oC); 5 min at 68oC.
M123456
kb
4.4-
2.0-
0.56-
CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 5
The Advantage-HF Kit ensures
high-fidelity amplification of ge-
nomic as well as cDNA tem-
plates. In Figure 3, the Advan-
tage HF Kit was used to amplify
a 2.1-kb fragment of the bovine
pancreas trypsin inhibitor gene
from different amounts of total
calf thymus DNA. The fragment
wasefficiently amplifiedfrom as
little as 25 ng of genomic DNA
(Lane 4).
Increaseelongationefficiency
for longer fragments
Tworeactionbuffersareincluded
inthe Advantage-HF Kit: theHF
Buffer and the original cDNA
Buffer. Use of the HF buffer de-
livers the highest possible fidel-
ity, as represented in Figure 1.
Fragments of up to ~2.5 kb can
be amplified under these condi-
tions. To amplify longer frag-
ments, some of the increase in
fidelity can be sacrificed to im-
prove elongation efficiency by
combining the HF and cDNA
buffers in varying proportions.
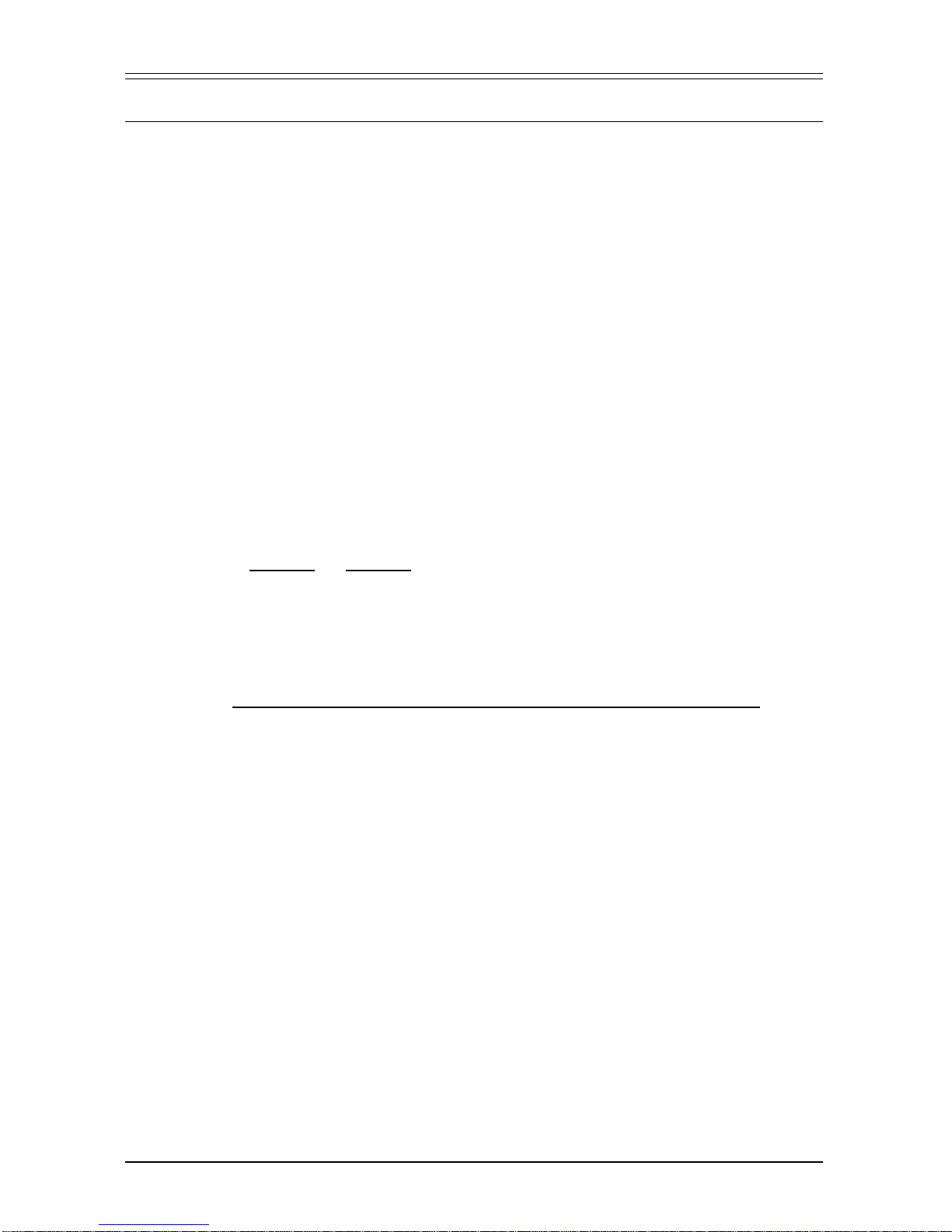
Figure 4, Panel A shows the
fidelity resulting from the use of
varying percentages of the HF
Buffer(seep.3fordescriptionof
assay). Panel B demonstrates
amplification of a 6.0-kb cDNA
fragmentusingtheindicatedcon-
centrations of HF Buffer. In this
example, the fragment is suc-
cessfully amplified in 80% HF
Buffer,conditionsallowinga~3-
fold increase in fidelity over the
cDNAbuffer.Optimalconditions
fortheamplificationofotherfrag-
ments should be determined in-
dividually.
I. Introduction
continued
Figure4. TheeffectofHFBufferconcentrationonPCR
fidelity (A) and amplification (B) of a 6.0-kb cDNA
fragment from Human Placenta cDNA. Size markers
are λ/
Hin
d III DNA.
Figure3.Advantage-HFamplificationfromgenomic
DNA. The indicated amounts of calf thymus DNA were
used to amplify a 2.1-kb fragment of the bovine pan-
creas trypsin inhibitor gene. M: λ/
Hin
d III DNA size
markers. Cycling parameters are the same as in Fig-
ure 2.
M 1 2345
ng: 100 75 50 25 0
B
200 40 60 80 100
0
0.5
1.0
1.5
2.0
HF Buffer %:
Mutants (%)
A
2.0-
4.4-

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
6 Version # PR76834 FAX:415-424-1064 or 800-424-1350
Automatic hot start with TaqStart Antibodies
Advantage-HF provides hot start PCR by including TaqStartTM Antibody in the
polymerase mix, eliminating the need for additional pipetting or handling steps.
"Hotstart" refers to anymethodfor assembling PCR reactionsthatkeeps one or
moreofthereactioncomponentsphysicallyorfunctionallyseparatefromtherest
of the components prior to the onset of thermal cycling. This prevents back-
grounddue to low-levelDNAsynthesis from nonspecificallyprimedsites prior to
the onset of thermal cycling. The advantages of hot start PCR have been
demonstrated in many different applications. However, only CLONTECH's
Advantage Polymerase Mixes provide automatic hot start.
TaqStart is a neutralizing monoclonal antibody directed against
Taq
DNA
polymerase.TaqStartrecognizesbothnative
Taq
andN-terminaldeletionssuch
as KlenTaq-1. When premixed with the appropriate polymerase, the antibody
blocks polymerase activity during the set-up of the PCR reactions at ambient
temperatures. Polymerase activity is restored at the onset of thermal cycling
becausethe antibody isdenaturedby temperatures greaterthan60°C.Theloss
of inhibition is complete and irreversible, so the polymerase regains its full
enzymatic activity for PCR.
TaqStart-mediated hot start PCR has been shown to significantly improve the
efficiency and specificity of DNA amplifications (Kellogg
et al
., 1994;
CLONTECHniques,
April 1994). Antibody-mediated hot start with TaqStart has
been proven to be at least as effective as manual hot start (d'Aquila
et al
., 1991)
or wax-bead-mediated hot start (Chou
et al
., 1991). In particular, TaqStart
reduces or eliminates nonspecific amplification products and primer-dimer
artifacts. In some cases, specific products can only be obtained by using
TaqStart.
A PCR system for every application
The Advantage cDNA and Genomic PCR Kits, containing the Advantage cDNA
and Genomic Polymerase Mixes, respectively, are designed for high-efficiency,
long-distance PCR amplification and are the foundation of the Advantage PCR
EnzymeSystems(
CLONTECHniques
,July1995;Barnes,1994).Theseversatile
kits are designed for high-performance amplification in the vast majority of PCR
applications(includingallof our PCR-based kits) and have firmly established the
benefits of PCR enzyme mixes containing hot start antibodies. Our other Advan-
tage-based kits have been developed for certain specialized applications. The
Advantage-GC cDNA and Genomic PCR Kits and Mixes (
CLONTECHniques
,
January 1997) combine the high efficiency of the Advantage system with a novel
reagent, GC-MeltTM, and an optimized buffer containing DMSO to permit amplifi-
cation of problematic GC-rich templates (up to 90% GC). As the newest addition
to the Advantage line, Advantage-HF allows a new level of fidelity, balanced by
the efficiency that is characteristic of Advantage.
I. Introduction
continued

CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 7
Advantage-HF PCR Kit (#K1909-1[100 rxns], 1909-y [10 rxns])
Store all components at –20°C. Enough reagents are supplied for 100 PCR
reactions of 50 µl each.
II. List of Components
10 rxns 100 rxns
•10 µl 100 µl 50X Advantage-HF Polymerase Mix
IncludesKlenTaq-1DNApolymeraseandTaqStartAntibody
(1.1 µg/µl) in the following storage buffer.
Concentration 1X
in 50X Component Concentration
50 % Glycerol 1.0 %
40 mM Tris-HCl (pH 7.5) 0.8 mM
50 mM KCl 1.0 mM
25 mM (NH4)2SO40.5 mM
1 mM EDTA 20 µM
5.0 mM β-mercaptoethanol 0.1 mM
0.25 % Thesit 0.005 %
Deep VentRTM is a minor component of the Advantage-HF
Polymerase Mix.
•60 µl 600 µl 10X HF PCR reaction buffer
•60 µl 600 µl 10X cDNA PCR reaction buffer
•60 µl 600 µl 10X HF dNTP mix
•400 µl 4 ml Purified Water (Millipore-purified)
•10 µl 100 µl Control DNA template
λDNA (0.2 ng/µl)
•10 µl 40 µl Control primer mix (10 µM each)
The sequences are:
5' primer 5'–TTGGTTGATCGTGGTGCAGAGAACGTTG–3'
3' primer 5'–GAGAAGGTCACGAATGAACCAGGCGATAA–3'

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
8 Version # PR76834 FAX:415-424-1064 or 800-424-1350
III. Additional Materials Required
The following reagents are required but not supplied.
• Mineral oil (We recommend Sigma®Cat. #M-3516.)
•0.5-ml PCR reaction tubes (We recommend Perkin-Elmer GeneAmpTM
0.5-ml reaction tubes [Cat. #N801-0737].)
•Thermal cycler (Perkin-Elmer DNA Thermal Cycler 480, 9600, or equivalent)
•Dedicated pipettors (1–2-µl, 1–10-µl, 1–20-µl, 20–200-µl, 200–1000-µl)
•PCR pipette tips suitable for the above pipettors and equipped with
aerosol-barrier filters. Do not autoclave pipette tips.
•DNA size markers (See Section IV.D)
•5X Stop/loading buffer (Sambrook
et al.
[1989] provides several
recipes.)

CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 9
PLEASE READ ENTIRE PROTOCOL BEFORE STARTING.
A. General Considerations
1. Thermal cycler
The guidelines for cycling parameters in this protocol have been
developed for a Perkin-Elmer DNA Thermal Cycler 480 or 9600 and
Perkin-Elmer GeneAmp 0.5-ml PCR reaction tubes. Newer cyclers
mayallowforshortercycletimesandeliminatetheneedtouseoil.
Theoptimalcyclingparametersmayvarywithdifferenttemplates,
primers, experimental protocols, tubes, and thermal cyclers.
Refer to the Troubleshooting Guide (Section V) for suggestions on
optimizing PCR conditions.
2. Primer design
PrimerdesignisthesinglelargestvariableinPCRapplicationsandthe
single most important factor in determining the success or failure of
PCR reactions.
Always check and recheck your primer design before
constructing or ordering primers.
CLONTECH offers PRIMER
PREMIER (#V1072-1, V1079-1), powerful, easy-to-use software that
ensures optimal primer design.
Length and G/C content: The Advantage-HF PCR Kit can be used in
a wide variety of PCR applications, and the constraints on primer
design will vary from one application to the next. In general, however,
primers should have a Tmof at least 70°C to achieve optimal results in
a two-step cycling program with a 68°C annealing/extension step.
Therefore, whenever possible, primers should be
at least
22 nucleotides
(nt) long (25–30-mers are preferred) and have a GC content of 45–60%.
3. Template quality
BecauseoftheexponentialnatureofPCRamplification,manyconven-
tional PCR applications work well with templates of average or even
low quality. However, the longer the target, the more important tem-
plate quality becomes. This is because the number of unnicked, full-
lengthtargetsdecreasesasthetargetlengthincreases,sopoorquality
DNA will have very few large, unnicked targets. Furthermore, some
depurination occurs when DNA is denatured during thermal cycling,
and this can lead to strand scission. Therefore, it is particularly
important to prepare high-quality, high molecular weight DNA when
amplifying large targets.
Templatequalityisalsoimportantwhenthehighestpossiblesensitivity
isneeded.Furthermore,incDNAapplicationssuchasRACEandother
RT-PCR protocols, incomplete reverse transcription can lead to an
absence of product, shorter than full-length products, or smearing.
For 5' and 3' RACE and general PCR from cDNA, you can ensure the
qualityofyourcDNAbyusingMarathon-ReadycDNAfromCLONTECH.
IV. Advantage-HF PCR Kit Protocol

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
10 Version # PR76834 FAX:415-424-1064 or 800-424-1350
IV. Advantage-HF PCR Kit
continued
4. Good PCR practices
a. Prepare reactions with dedicated pipettors in a dedicated work space
DuetothetremendousamplificationpowerofPCR,minuteamounts
of contaminating DNA can produce nonspecific amplification; in
some instances, contaminants can cause DNA bands even in the
absence of added template DNA. We recommend that you set up
your PCR reactions in a dedicated lab area or noncirculating
containment hood and use dedicated pipettors, PCR pipette tips
with hydrophobic filters, and dedicated solutions.
Perform post-
PCR analysis in a separate area with a separate set of pipettors.
b. Pipetting
Because of the small volumes used in PCR experiments and the
potential for tube-to-tube variation, careful pipetting technique is
extremelyimportant.Alwaysbesurethatnoextrasolutionisonthe
outside of the pipette tip before transfer. When adding solution to a
tube, immerse the tip into the reaction mixture, deliver the solution,
and rinse the pipette tip by pipetting up and down several times.
c. Use a Master Mix
Using a Master Mix greatly reduces tube-to-tube variation. There-
fore,useaMasterMixwheneveryousetupmultiplePCRreactions.
Ifmultipletemplatesarebeingtestedwiththesameprimers,include
the primers in the Master Mix. If one template is being tested with
multiple primer sets, include the template in the Master Mix. For
several sets of parallel samples, assemble multiple master mixes
(e.g., each with a different set of primers).
The Master Mix should
be thoroughly mixed before use (i.e., vortexed without bubbling).
d. Include positive and negative controls (i.e., H2O instead of DNA
template)
5. Touchdown PCR
We have found that "touchdown" PCR significantly improves the
specificity of many PCR reactions in a wide variety of applications
(Section V.B.; Don
et al.
, 1991; Roux, 1995). Briefly, touchdown PCR
involves using an annealing/extension temperature that is several
degrees(typically3–10°C)
higher
thantheTmoftheprimersduringthe
initial PCR cycles (typically 5–10). The annealing/extension tempera-
ture is then reduced to the primer Tmfor the remaining PCR cycles.
6. TaqStart Antibody provides automatic hot start PCR
Donot use a manual hot start or wax-bead-based hot start when using
Advantage-HF. As discussed in the Introduction, hot start is automatic
with Advantage-HF because the enzyme mix already contains TaqStart
Antibody.
CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 11
7. Use of additives
TaqStart Antibody binds KlenTaq-1 DNA polymerase with high affinity
under the conditions described in this protocol. The addition of 2–5%
DMSOwillnotinterferewithTaqStartfunctionandmayimproveresults
in some instances (see Section V.A.). However, the addition of
formamideor other cosolvents maydisruptTaqStart function. Further-
more, excessive glycerol, solutes (e.g., salts), pH extremes, or other
deviationsfrom the recommended reaction conditions may reduce the
effectiveness of the antibody and/or DNA polymerases.
B. Control PCR Reactions
The following PCR reactions can be performed in parallel with your
experiments as controls to ensure that the Advantage-HF Kit is working
properly. A positive control template and primers are provided in the kit.
1. Place all components on ice and allow to thaw completely. Mix each
component thoroughly before use.
2. Combine the following reagents in a 0.5-ml PCR tube.
Positive Negative
Control Control
32 µl37µl Purified H2O
5µl5µl 10X HF PCR reaction buffer
5µl --- Control DNA template (~0.2 ng/µl)
2µl2µl Control primer mix (10 µM ea.)
5µl5µl 10X HF dNTP mix
1µl1µl 50X Advantage-HF Polymerase Mix
50 µl50µl Total
3. Mix well and spin the tube briefly to collect all the liquid in the bottom
of the tube.
4. Add 1–2 drops of mineral oil to prevent evaporation during cycling and
cap firmly. A good "capping" of mineral oil should have a well-defined
meniscus between the two phases.
5. Commence thermal cycling. If using a Perkin-Elmer DNA Thermal
Cycler Model 480 or 9600, use the parameters described in Section C
below.20–22cycleswitha4-minannealing/extensiontimeissufficient
for amplification of the positive control template provided in the kit.
6. Transfer a 5-µl sample of your PCR reaction to a fresh tube and add
1 µl of 5X stop/loading buffer. Analyze your sample(s), along with
suitableDNAsizemarkers,byelectrophoresisona0.8–1.2%agarose/
ethidium bromide gel.
Expected results: If you are using the positive control reagents provided
inthekit,thereactionshouldproduceasinglemajorbandof2kb.Nobands
should be generated in the negative (i.e., no DNA template) control.
IV. Advantage-HF PCR Kit
continued

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
12 Version # PR76834 FAX:415-424-1064 or 800-424-1350
IV. Advantage-HF PCR Kit
continued
C. Recommended Cycling Parameters
Use the following guidelines when setting up your initial experiments with
the Advantage-HF Polymerase Mix. These are general guidelines—the
optimalparametersmayvarywithdifferentthermalcyclersandwilldepend
on your particular primers and templates, and on other experimental
variables. Note: When using the Advantage-HF Kit with products such as
CLONTECH's MarathonTM cDNA Amplification Kit, Marathon-ReadyTM
cDNAs, or the DeltaTM Differential Display, use the parameters recom-
mended in the protocol for that kit.
Cycle Parameters
(PE 480) (PE 9600)
• 94°C for 1 min • 94°C for 15 sec
• 25–35 cycles a • 25–35 cycles a
94°C 30 sec b94°C 5–15 sec b
68°C 4 min c68°C 4 min c
• 68°C for 3 min d• 68°C for 3 min d
• Soak at 15°C • Soak at 15°C
a25 cycles for multiple-copy genes or medium-to-high abundance cDNAs; 30–35
cycles for single- or low-copy-number genes or rare cDNAs. For most applications,
weprefer two-stepcycles (denaturation atT1followedby annealingand extensionat
T2)overthree-step cycles(denaturationatT1followedbyannealingat T2followedby
extension at T3). Three-step cycles will be necessary when the Tmof the primers is
less than 60–65°C and in certain special protocols (such as Delta Differential Display).
b Use the shortest possible denaturation time. Exposure of DNA to high temperatures
causes some depurination of single-stranded DNA during denaturation, which
eventually leads to strand scission. High temperature also leads to gradual loss of
enzyme activity.
cUse the highest possible annealing/extension temperature. See Note a. Shorter
targets may be amplified using shorter extension times.
Some researchers prefer to use an annealing/extension time equal to the expected
target size plus two minutes.
dOptional: This final extension may reduce background in some cases.
CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 13
IV. Advantage-HF PCR Kit
continued
D. Amplification of Longer Fragments with the Advantage Buffer
The Advantage HF Kit is provided with two buffers—the HF Buffer and the
standard cDNA Buffer. When used with the HF Buffer, this kit delivers the
highestpossiblefidelity.Fragmentsofupto~2.5kbcanbeamplifiedunder
these conditions. To amplify longer fragments, some of the increase in
fidelity can be sacrificed to improve elongation efficiency by combining the
HF and cDNA buffers in varying proportions (see Figure 4). To amplify
longer fragments, we recommend replacing the smallest amount of HF
Buffer that allows satisfactory amplification. For example, the 6-kb frag-
ment in Figure 4 can be amplified from a cDNA library in a 50-µl PCR
reaction containing 4µl of 10X HF Buffer (80% final) and 1 µl of 10X cDNA
Buffer.Optimalconditionsfortheamplificationofotherfragmentsshouldbe
determined individually. We recommend initially trying HF Buffer concen-
trations in the 50–100% range for fragments up to 10 kb.
E. Recommendations for Electrophoresis
We recommend that you transfer a 5-µl sample of your PCR reaction to a
fresh tube and add 1 µl of 5X stop/loading buffer. (The remaining 45 µl of
the reaction mixture can be subjected to further cycling if you do not see a
product.) Analyze your sample(s), along with suitable DNA size markers,
by electrophoresis on a suitable agarose gel containing 0.1–0.5 µg/ml
ethidiumbromide.Thepercentage agaroseandtheDNAsizemarkersyou
choosewilldependontheexpectedrangeofinsertsizes.Youmaywishto
refer to the following general guidelines before assembling your gel.
Recommendations for agarose gels:
Expected Recommended Recommended
insert size range % agarose DNA size markers
0.3–1.5 kb 1.5 φX174/
Hae
III
0.5–10 kb 1.2 1-kb DNA ladder
>5 kb 0.8 λ/
Hin
d III

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
14 Version # PR76834 FAX:415-424-1064 or 800-424-1350
The following general guidelines apply to most PCR reactions. However, no
attempt has been made to address troubleshooting for all of the many applica-
tions for which the Advantage-HF Kit can be used. When using the kit with
another CLONTECH product, additional, application-specific troubleshooting
information can be found in the relevant User Manual.
A. No product observed
PCR component Use a checklist when assembling reactions. Always
missing or degraded perform a positive control to ensure that each com-
ponent is functional. If the positive control does not
work, repeat the positive control only. If the positive
control still does not work, repeat again replacing
individual components to identify the faulty reagent.
Too few cycles Increasethenumberofcycles(3–5additionalcycles
at a time).
Annealing temp. Decrease the annealing temperature in increments
too high of 2–4°C.
Suboptimal primer Redesign your primer(s) after confirming the accu-
design racy of the sequence information. If the original
primer(s) was less than 22 nt long, try using a longer
primer. If the original primer(s) had a GC content of
less than 45%, try to design a primer with a GC
content of 45–60%.
Not enough Repeat PCR using a higher concentration of DNA
template (after trying more cycles).
Poor template Check template integrity by electrophoresis on a
quality standard TBE-agarose gel. If necessary, repurify
yourtemplateusingmethodsthatminimizeshearing
and nicking.
Denaturation temp. Optimize denaturation temperature bydecreasingor
too high or low increasing it in 1°C increments. (A denaturation
temperature that is too high can lead to degradation
ofthetemplate,especiallyforlongtargetsequences.)
Denaturation time Optimize denaturation timeby decreasingorincreas-
too long or too short ingit in 10-sec increments. (A denaturation time that
is too long can lead to degradation of the template,
especially for long target sequences.)
Extension time too (Especially with longer templates) Increase the
short extension time in 1-min increments.
V. Troubleshooting Guide

CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 15
Too little enzyme Advantage-HFPolymeraseMixis50Xformostappli-
cations. Therefore, try to optimize the cycle param-
eters as described above before increasing the en-
zyme concentration. In rare cases, the yields can be
improved by increasing the concentration of the
enzyme mix. However, increasing the concentration
>2X is likely to lead to higher background levels.
[Mg2+] is too low KlenTaq-1 DNA polymerase has a broader Mg2+
optimum than native
Taq
DNA polymerase (i.e., it
performs well over a wider range of [Mg2+] with no
loss of efficiency.) Therefore, as long as you use the
buffers included in the kit, it is unlikely that a lack of
productisduetoproblemswiththeMg++concentration.
However, if the concentration of EDTA in the cDNA
sampleismorethan5mM,thiscanreducetheeffective
concentrationofMgtobelowaminimumlevel.Increas-
ing the concentration of Mg2+ can result in lower
fidelity.
[dNTPs] not optimal The Advantage-HF PCR Kit contains a carefully
balanced mixture of the 4 dNTPs. Replacement of
thismixturewithastandarddNTPmix(200µMeach)
may improve the DNA yield, but may also result in
lower fidelity.
Difficult target Some targets are inherently difficult to amplify. In
mostcases, this is due to unusually high GC content
and/orsecondarystructure.The Advantage-GCKits
are recommended in these situations.
B. Multiple products
Too many cycles Reducing the cycle number may eliminate nonspe-
cific bands.
Annealing temp. Increase the annealing/extension temperature in
too low increments of 2–3°C.
Suboptimal primer Redesign your primer(s) after confirming the accu-
design racy of the sequence information. If the original
primer(s) was less than 22 nt long, try using a longer
primer. If the original primer(s) had a GC content of
less than 45%, try to design a primer with a GC
content of 45–60%.
V. Troubleshooting Guide
continued

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
16 Version # PR76834 FAX:415-424-1064 or 800-424-1350
Touchdown PCR "Touchdown" PCR significantly improves the speci-
needed ficity of many PCR reactions in various applications
(Don
et al.
, 1991; Roux, 1995). Touchdown PCR
involves using an annealing/extension temperature
that is several degrees
higher
than the Tmof the
primersduringtheinitialPCRcycles.Theannealing/
extension temperature is then reduced to the primer
TmfortheremainingPCRcycles.Thechangecanbe
performed either in a single step or in increments
over several cycles.
Contamination See Section D below.
C. Products are smeared
Too many cycles Reduce the cycle number by 3–5 to see if non-
specific bands go away.
Denaturation temp. Tryincreasingthedenaturationtemperatureinincre-
too low ments of 1°C.
Extension time Decreasetheextensiontime in1–2-min increments.
too long
Poor template Checktemplate integrity by electrophoresis on a de-
quality denaturing agarose gel. Repurify your template if
necessary.
Touchdown PCR See "Touchdown PCR needed" under previous
needed section.
Too much enzyme Advantage-HFPolymeraseMixis50Xformostappli-
cations; however, a 1X final concentration of the
enzyme mix may be too high for some applications.
If smearing is observed, first try optimizing the cycle
parameters as described above, then try reducing
the enzyme concentration to 0.5–0.2X.
[Mg2+] is too high KlenTaq-1 DNA polymerase has a broader Mg2+
optimum than native
Taq
DNA polymerase (i.e., it
performs well over a wider range of [Mg2+] with no
loss of efficiency.) Therefore, as long as you have
used the buffers supplied in the kit, it is unlikely that
smearing is due to problems with the Mg2+
concentration. Altering the concentration ofMg++ can
result in lower fidelity.
Too much template Try a lower concentration of DNA template in the
PCR reaction.
Contamination See Section D below.
V. Troubleshooting Guide
continued

CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 17
D. Dealing with contamination
Contaminationmostoftenresultsinextrabandsorsmearing.Itisimportant
to include an H2O control (i.e., a control using H2O instead of the DNA
template) in every PCR experiment to determine if the PCR reagents,
pipettorsorPCRreactiontubesarecontaminatedwithpreviouslyamplified
targets.
If possible, set up the PCR reaction and perform the post-PCR analysis in
separate laboratory areas with separate sets of pipettors.
Laboratory benches and pipettor shafts can be decontaminated by
depurination. Wipe surfaces with 1N HCl followed by 1N NaOH. Then
neutralize with a neutral buffer (e.g., Tris or PBS) and rinse with H2O.
Itisadvisabletouseoneofthecommerciallyavailableaerosol-freepipette
tips.
There is an enzymatic method for destroying PCR product carryover
(Longo
et al.
, 1990). It involves incorporation of dUTP into the PCR
products and subsequent hydrolysis with uracil-N-glycosylase (UNG).
When performing PCR directly on phage plaques or bacterial colonies,
failuretoisolatesingleplaquesorcolonieswillalsoproducemultiplebands.
V. Troubleshooting Guide
continued

CLONTECH Laboratories, Inc.
page Protocol # PT3139-1 Technical Support TEL:415-424-8222 or 800-662-CLON
18 Version # PR76834 FAX:415-424-1064 or 800-424-1350
VI. References
Advantage-GC PCR Kits (January 1997)
CLONTECHniques
XII(1):2–3.
Barnes, W. M. (1994) PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ
bacteriophage templates.
Proc. Natl. Acad. Sci. USA
91:2216–2220.
Cariello, N. F., Swenberg, J. A. & Skopek, T. R. (1991) Fidelity of
Thermococcus litoralis
DNA
polymerase (Vent) in PCR determined by denaturing gradient gel electrophoresis.
Nucleic Acids
Res.
19(15):4193–4198.
Cheng, S., Fockler, C., Barnes, W. M. & Higuchi, R. (1994) Effective amplification of long targets
from cloned inserts and human genomic DNA.
Proc. Natl. Acad. Sci. USA
91:5695–5699.
Chou,Q.,Russell,M.,Birch,D.,Raymond,J.&Bloch,W.(1992)Preventionofpre-PCRmispriming
and primer dimerization improves low-copy-number amplifications.
Nucleic Acids Res.
20:1717–
1723.
d'Aquila, R. T., Bechtel, L. J., Videler, J. A., Eron, J. J., Gorczyca, P. & Kaplan, J. C. (1991)
Maximizing sensitivity and specificity of PCR by preamplification heating.
Nucleic Acids Res.
19:3749.
Don, R. H., Cox, P. T., Wainwright, B. J., Baker, K. & Mattick, J. S. (1991) 'Touchdown' PCR to
circumvent spurious priming during gene amplification.
Nucleic Acids Res.
19:4008.
Frey, B. & Suppmann, B. (1995) Demonstration of the ExpandTM PCR system's greater fidelity and
higher yields with a
lacI
-based PCR fidelity assay.
Biochemica
2:8–9.
Kellogg,D.E.,Rybalkin,I.,Chen,S.,Mukhamedova,N.,Vlasik,T.,Siebert,P.&Chenchik,A.(1994)
TaqStart Antibody: Hotstart PCR facilitated by a neutralizing monoclonal antibody directed against
Taq
DNA polymerase.
BioTechniques
16:1134–1137.
Ling, L. L., Keohavong, P., Dias, C. & Thilly, W. G. (1991) Optimization of the polymerase chain
reaction with regard to fidelity: modified T7,
Taq
, and Vent DNA polymerases.
PCR Methods Appl.
1:63–69.
Longo,M.C.,Berninger,M.S.&Hartley,J.L.(1990)UseofuracilDNAglycosylasetocontrolcarry-
over contamination in polymerase chain reactions.
Gene
93:3749.
Mo,J. Y.,Maki, H.& Sekiguchi, M.(1991) Mutationalspecificity of thednaE173 mutatorassociated
withadefectinthecatalyticsubunitofDNApolymeraseIIIof
Escherichiacoli
.
J.Mol.Biol.
222:925–
936.
Nelson, K., Brannan, J. & Kretz, K. (1995) The fidelity of TaqPlusTM DNA Polymerase in PCR.
Strategies Mol. Biol.
8:24–25.
Roux, K. H. (1995) Optimization and troubleshooting in PCR.
PCR Methods Appl
. 4:5185–5194.
Sambrook,J., Fritsch, E.F. & Maniatis,T.(1989)
MolecularCloning: A Laboratory Manual, Second
Edition
(Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).

CLONTECH Laboratories, Inc.
TEL:415-424-8222 or 800-662-CLON Technical Support Protocol # PT3139-1 page
FAX:415-424-1064 or 800-424-1350 Version # PR76834 19
VII. Related Products
Product Cat. #
• Advantage®cDNA PCR Kit #K1905-1, -y
• Advantage®cDNA Polymerase Mix #8417-1
• Advantage®Genomic PCR Kit #K1906-1, -y
• Advantage®Genomic Polymerase Mix #8418-1
• Advantage®-GC cDNA PCR Kit #K1907-1, -y
• Advantage®-GC cDNA Polymerase Mix #8419-1
• Advantage®-GC Genomic PCR Kit #K1908-1, -y
• Advantage®-GC Genomic Polymerase Mix #8420-1
• Advantage®UltraPure dNTPs (many)
Other Related Products
• TaqStartTM Antibody #5400-1, -2
• TthStartTM Antibody #5401-1
• PRIMER PREMIER many
• Poly A+RNA many
• Multiple Tissue Northern (MTNTM) Blots many
• UltraPure PCR Deoxynucleotide Mix #4700-1

Advantage®, Advantage®-GC, Advantage®-HF,
DeltaTM, Marathon-ReadyTM, TaqStartTM, and TthStartTM
are trademarks of CLONTECH Laboratories, Inc.
GeneAmpTM is a trademark of Hoffmann-La Roche, Inc.
Deep VentRTM is a trademark of New England Biolabs,
Inc.
© 1997, CLONTECH Laboratories, Inc. All rights
reserved.
Table of contents
Popular Laboratory Equipment manuals by other brands

Turner Designs
Turner Designs TD-700 operating manual

Excillum
Excillum MetalJet D2+ operating manual

Hettich
Hettich ROTINA 35 R Repair instructions

SCP SCIENCE
SCP SCIENCE DigiPREP HP user manual

PASCO
PASCO OS-9255A instruction manual

Thermo Scientific
Thermo Scientific Sorvall Legend X1R instruction manual











