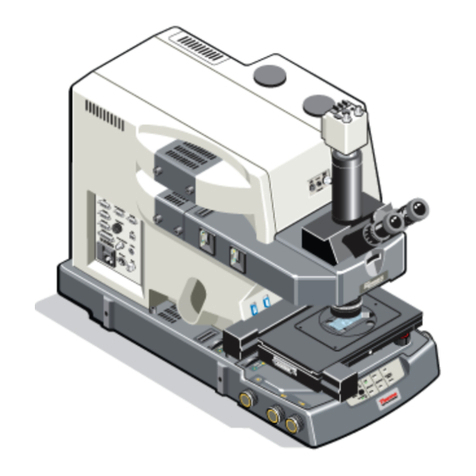
Stereology with the Optical Fractionator Workow
Tissue Preparation
The optical fractionator requires fairly
thick tissue. Thicker tissue allows one to
distinguish between cells that are on top
of each other while focusing through
the tissue making it easier to see the
cells clearly as they start to come into
focus. Optimal thickness for this method
is 20 microns after accounting for shrink-
age from tissue processing.
Options here are:
1. Cut thicker sections.
2. Dry the tissue as little as possible.
3. Avoid unnecessary alcohol steps.
Collect serial tissue sections without
losing any sections along the way. The
Optical Fractionator requires a series of
sections, such as every 3rd section. The
interval will depend on the size of the
structure and how many cells the struc-
ture contains; this can be determined
with a pilot study.
Do a Test Run or Pilot Study
A pilot study will help decipher the best
parameters to use for each experiment
including counting frame size, grid size,
and serial section interval. Due to vari-
ability across animals, a pilot study may
be required on one animal from each
group within the experiment.
With the Optical Fractionator the target
is to count no less than 200 cells per
animal. This can be achieved by count-
ing 10 sections with 10 sampling sites
per section and on average 2 cells per
sampling site. Make sure the Coecient
of Error (CE) is below 0.1. The best way
to approach a pilot study is to err on
the side of over-counting. Once cells
counts have been completed and the
CE has been calculated for the test
animal, you can decide whether or not
to adjust the parameters.
Once the results of a pilot study, have
been reviewed, adjustments to the pa-
rameters can be made in order to count
more cells or fewer cells as necessary.
Ways to DECREASE the number of
cells:
1. Make the counting frame smaller.
2. Decrease the grid size creating fewer
sampling sites.
3. Increase the section interval, for
example, counting every 4th section
instead of every 3rd.
Ways to INCREASE the number of
cells you count:
1. Make the counting frame bigger.
2. Make the grid size smaller so that
you visit more sites.
3. Decrease the section interval, for
example, counting every 3rd section
instead of every 4th.
Once good parameters for an experi-
ment have been determined, they can
be saved and quickly pulled up in the
workow for subsequent animals. Do-
ing a pilot study will ensure that quality
data is generated by obtaining the best
estimates of total cell number.
Step 1: Set up the subject
1. Number of sections to count: De-
fault is 1; new sections can be added as
you go.
2. Section’s cut thickness: How thick the
tissue was cut at.
3. Section Evaluation Interval: This is
the interval between the sections to
count. Every other section =2, every 3rd
=3 ...
4. Starting Section Number: Default;
start counting at section 1. The rst
section is dened as the rst section that
displays the desired morphology.
5. Z-value of First Section: This will
always be 0.00 at section 1. If starting on
a section other than one, it will be an in-
terval of the cut thickness. For example
if tissue was cut at 50 microns and you
are starting on section one, the Z value
=0.00, if starting on section 2 the Z value
=50, if starting on section 3, the Z value
= 100, and so on.
Step 2: Set microscope to Low Magni-
cation
Choose a low power objective (2.5x, 5x
or 10x) best tted for tracing a contour
around your region of interest (ROI).
Step 3: Trace your Region of Interest
(ROI)
Select a contour to use and then trace.
Use this same contour choice for the
same region of interest in subsequent
sections. More than one contour can
be traced to count dierent regions in a
structure or dierent structures. Con-
tours can be named uniquely.
Hint: Name contours after your area of
interest so they can quickly be selected
in the Display menu. Click on Display Set-
tings and then the contour tab. Click on
the contour to rename. The change will
be reected in the workow.
Step 3 is a good place to edit a contour.
Go to the Edit menu and click on Select
Objects. Then click on the contour and
it will be highlighted with white squares.
Once it is highlighted, right click for
various editing tools. Remember to right
click and Exit the Selection Tool once
editing is complete.
Step 4: Set Microscope to High Magni-
cation
Select a high power objective that will
be good for counting (40x-oil, 63x-oil or
These Parameters should be set so
that you end up counting at least
200 cells and/or a CE of less than 0.1
is achieved without spending time
counting more cells than you need to.
Also, do not change parameters for
every section within an animal! All
parameters must be kept constant
throughout all the sections of the
animal for the calculations to be
valid. It is best to keep the same pa-
rameters for each group of animals
as well.
NOTE: counting frame size, grid size, and serial section interval
Information below is derived from MBF Bioscience correspondence