________________________________________________________________
___
10 English Issue 02 - 07/2001
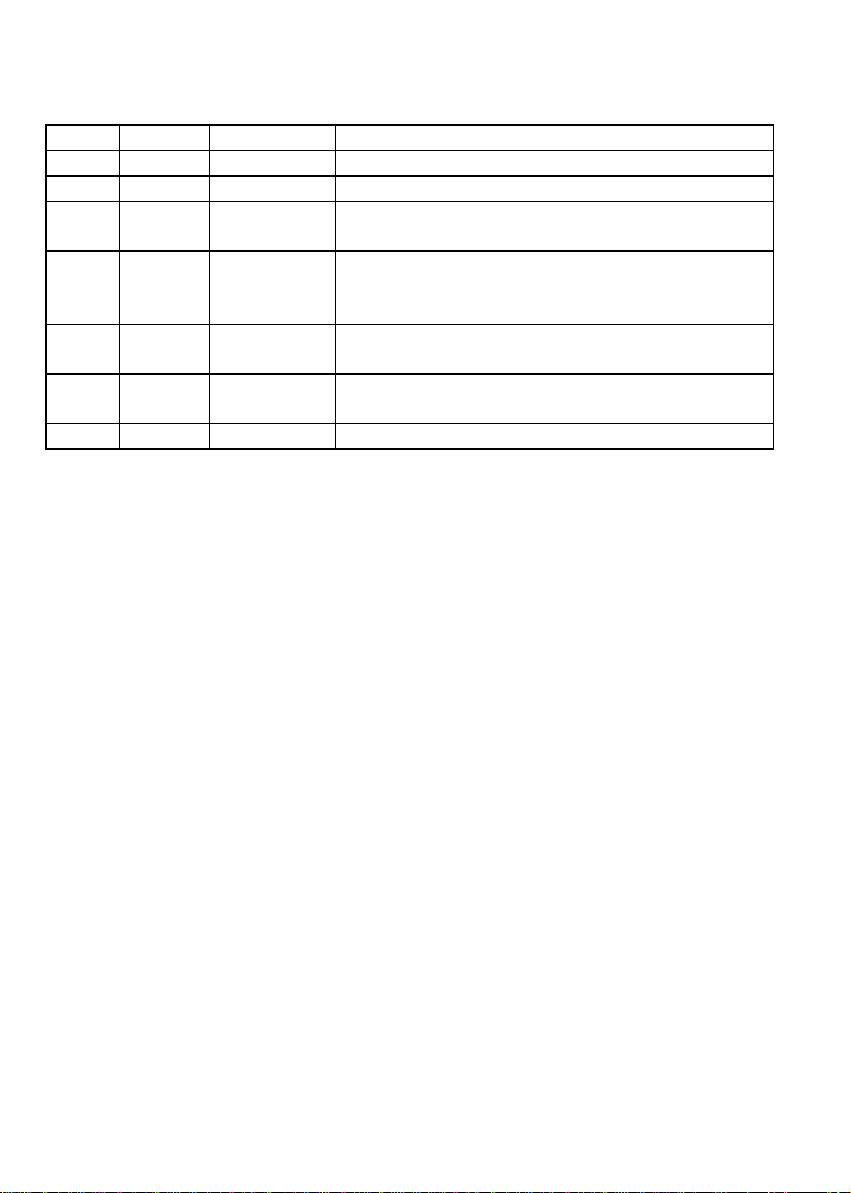
Nucleic
Mode Factor A260/A280 Use
DNA 50 ng/µl 1.8 DNA quantification and purity checking
RNA 40 ng/µl 2.0 RNA quantification and purity checking
Oligo 33 ng/µl Sequence
dependent Oligonucleotide quantification and purity checking
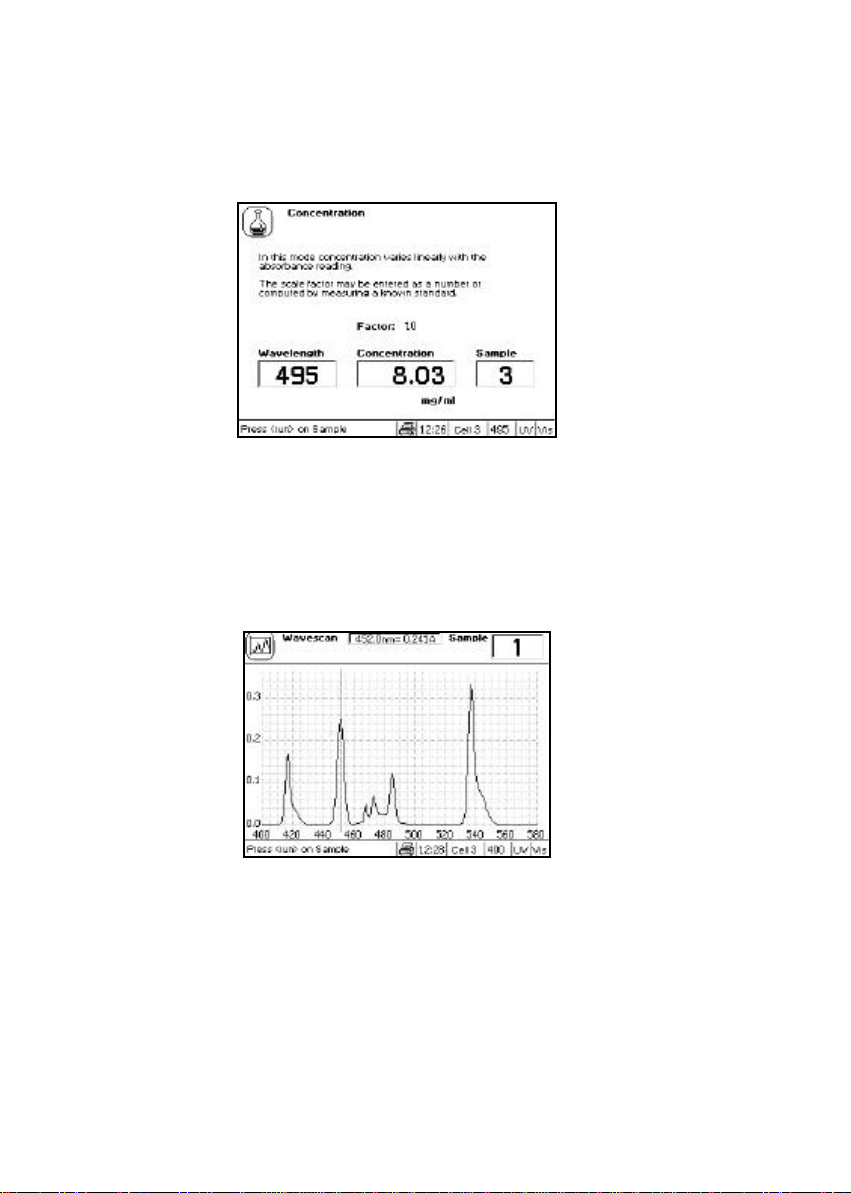
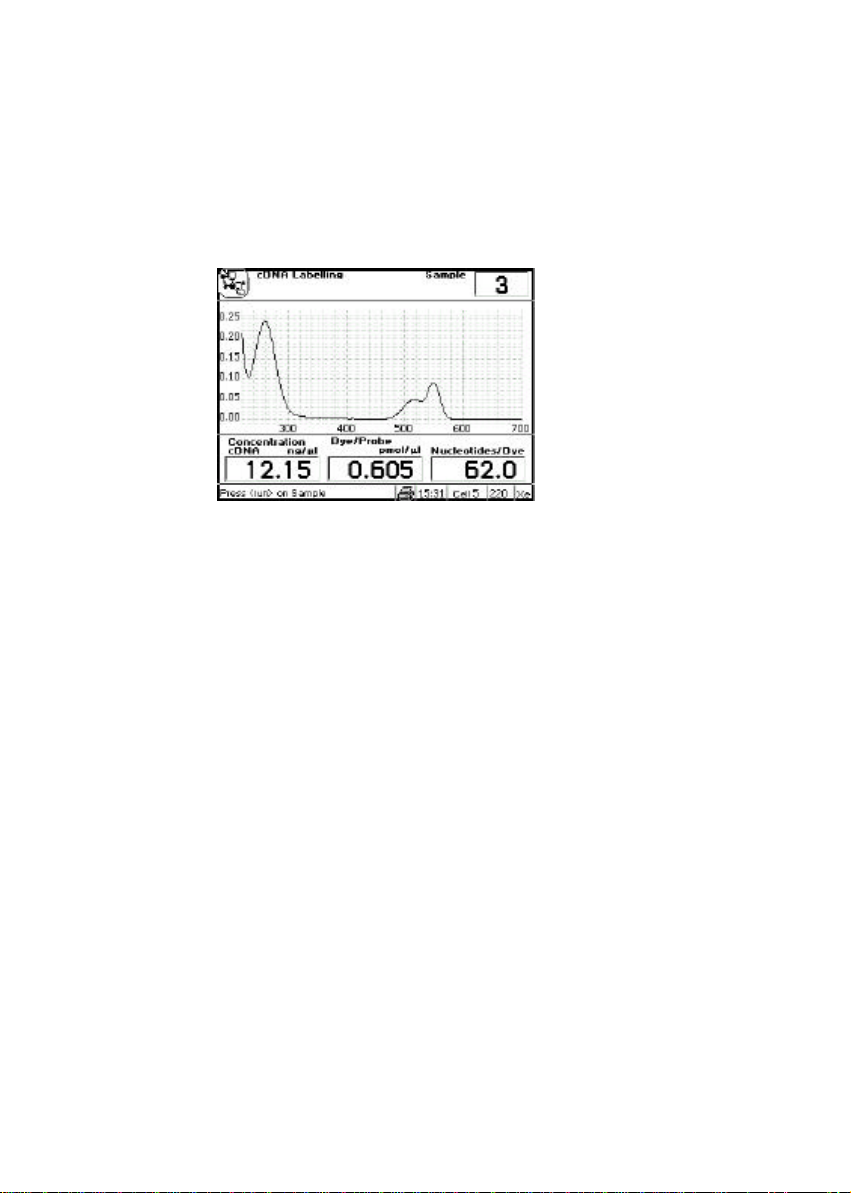
cDNA
label 50 ng/µl Fluorescent cDNA and PCR probe quantification
for micro-arrays andin-situ hybridisation studies,
respectively
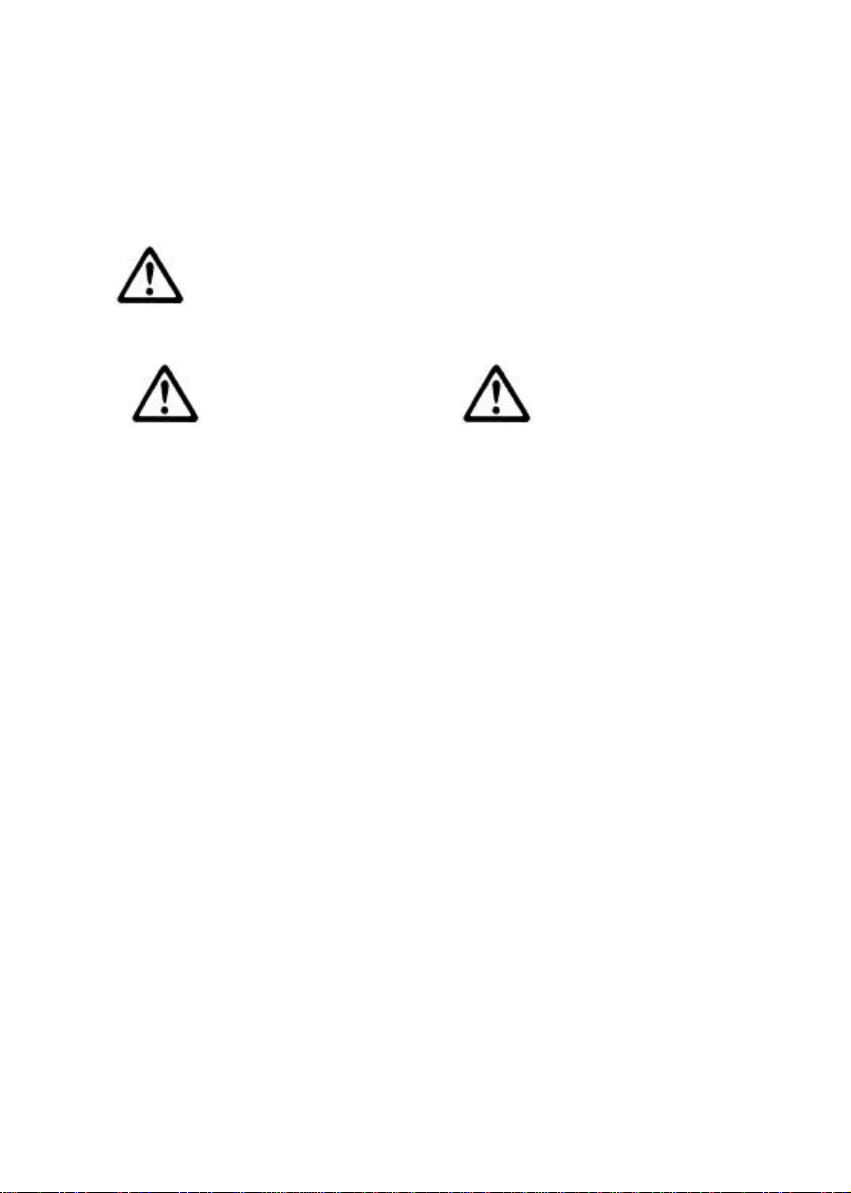
Scan - - Spectrum of samples, as well as quantification and
purity check of selected wavelengths
Tm - - Calculate theoretical Tm for a nucleotide base
sequence
Info - - Information relating to nucleic acids
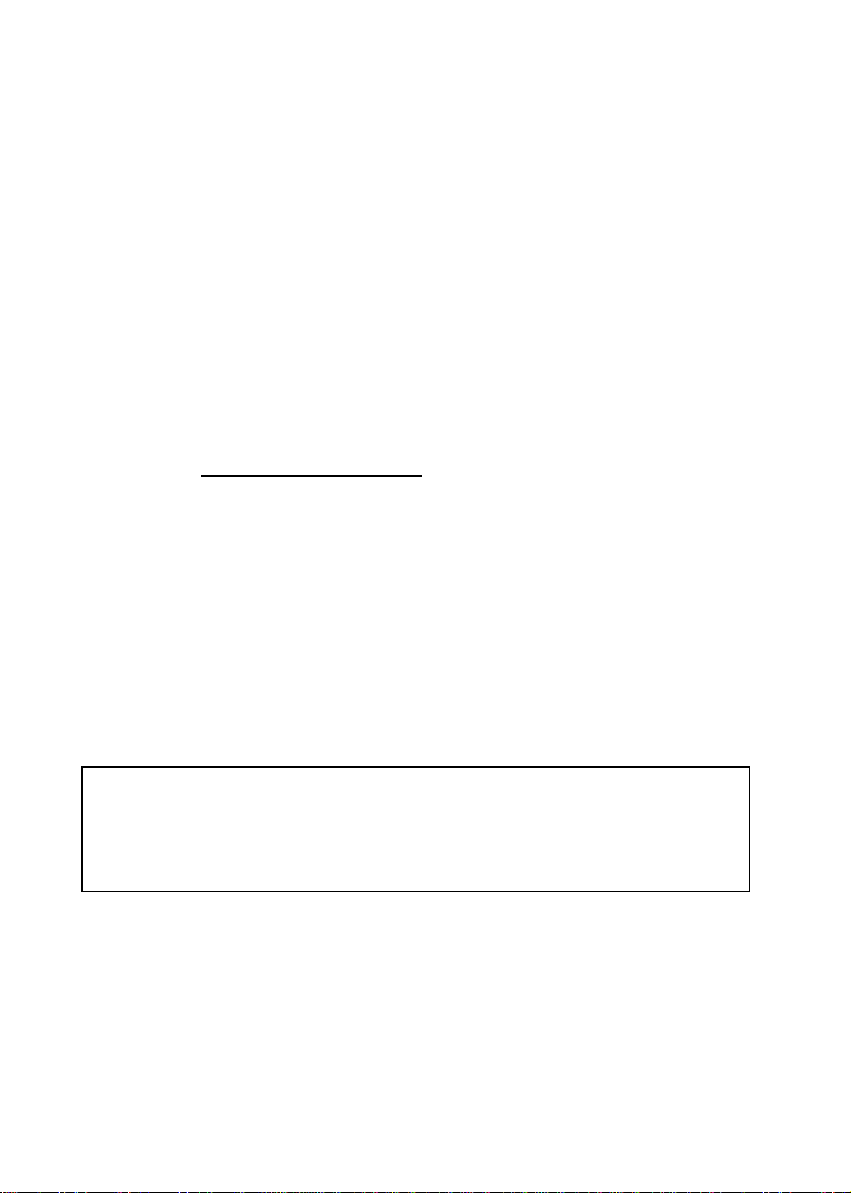
Nucleic acids can be quantified at 260 nm because it is well established that a
solution of DNA or RNA with an optical density of 1.0 has a concentration of 50 or
40 µg/ml, respectively, in a 10mm pathlength cell. Oligonucleotides, as a rule of
thumb, have a corresponding factor of 33 µg/ml, although this does vary with base
composition.
Concentration = Abs260 * Factor
Extracting nucleic acids from cells is accompanied by protein, and extensive
purification is required to separate the protein impurity. The 260/280 ratio gives an
indication of purity; it is only this, however, and not a definitive assessment. Pure
DNA and RNA preparations have expected ratios of ≥1.8 and≥2.0, respectively;
deviations from this indicate the presence of protein impurity in the sample, but care
must be taken in interpretation of results. An elevated absorbance at 230 nm can
indicate the presence of impurities as well; 230 nm is near the absorbance maximum of
peptide bonds and also indicates buffer contamination since Tris, EDTA and other
buffer salts absorb at this wavelength. When measuring RNA samples, the 260/230
ratio should be > 2.0; a ratio lower than this is generally indicative of contamination
with guanidinium thiocyanate, a reagent commonly used in RNA purification and
which absorbs over the 230 – 260 nm range. A wavelength scan of a sample can also
be obtained for visual inspection of integrity over the range 200 – 350 nm.
Absorbance ratio = Abs260 / Abs280
cDNA and PCR tagged with fluorescent probes can be scanned up to 850 nm so that
both peaks can be used to monitor labelling efficiency.