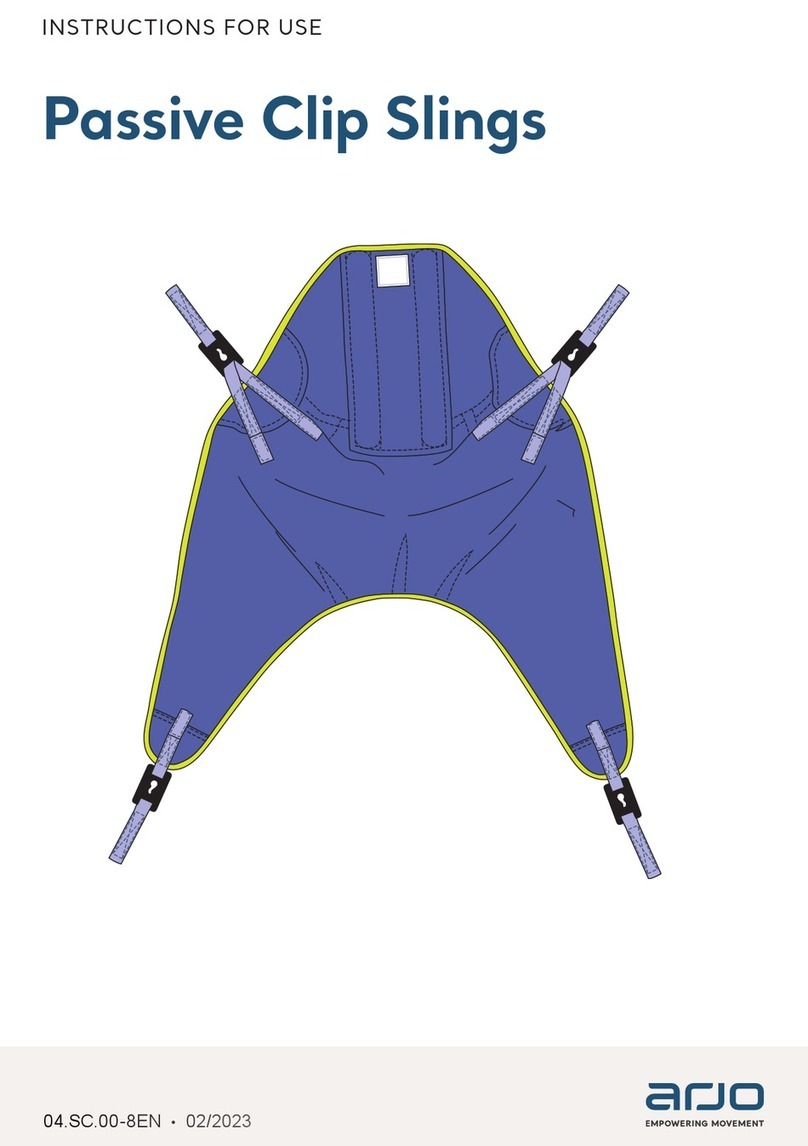
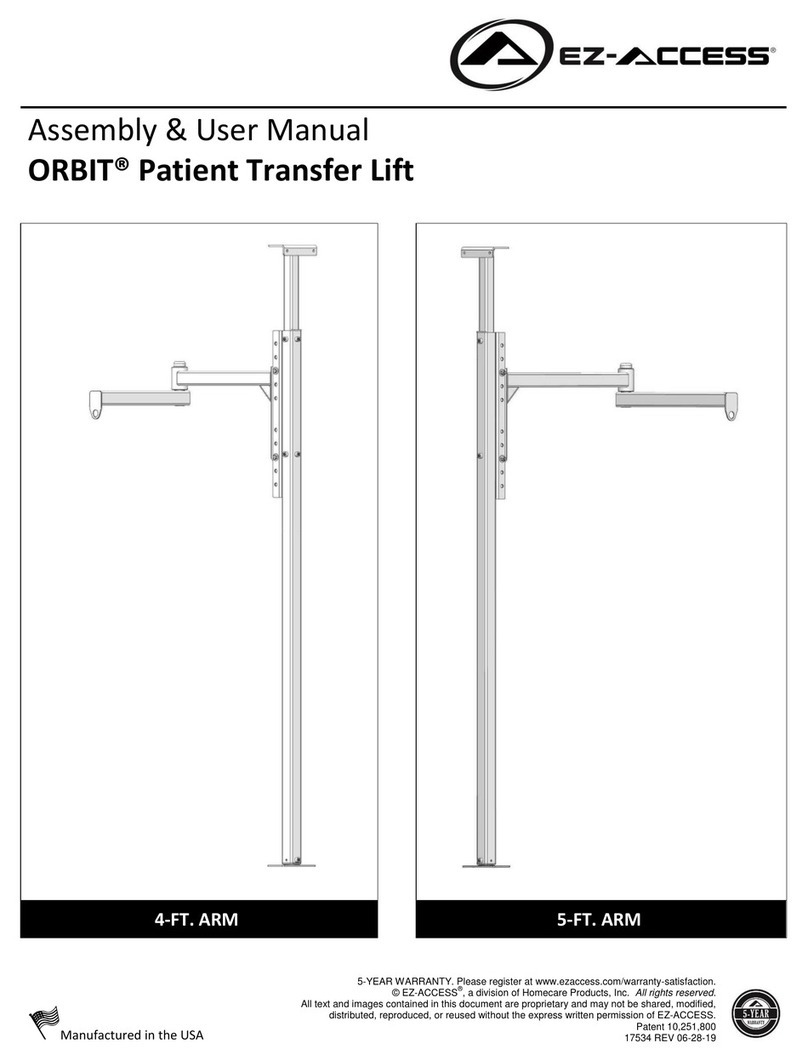
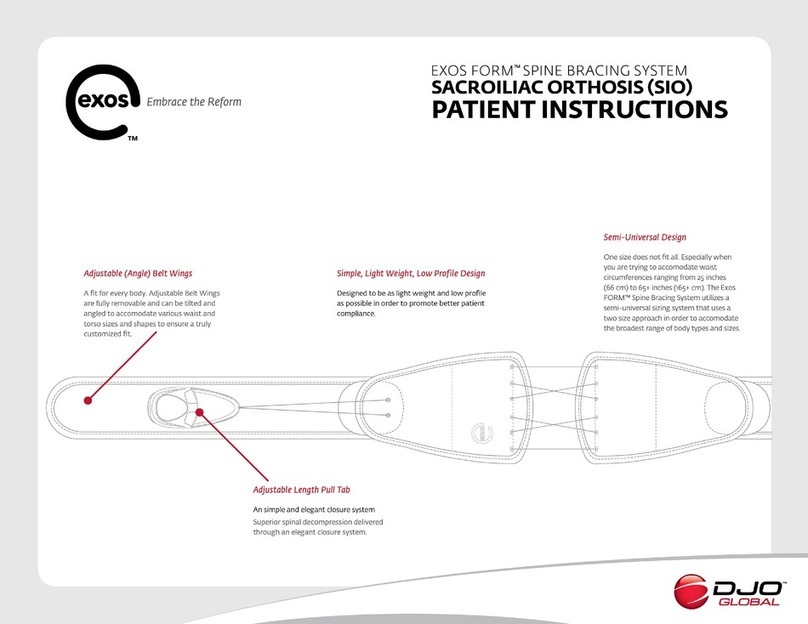
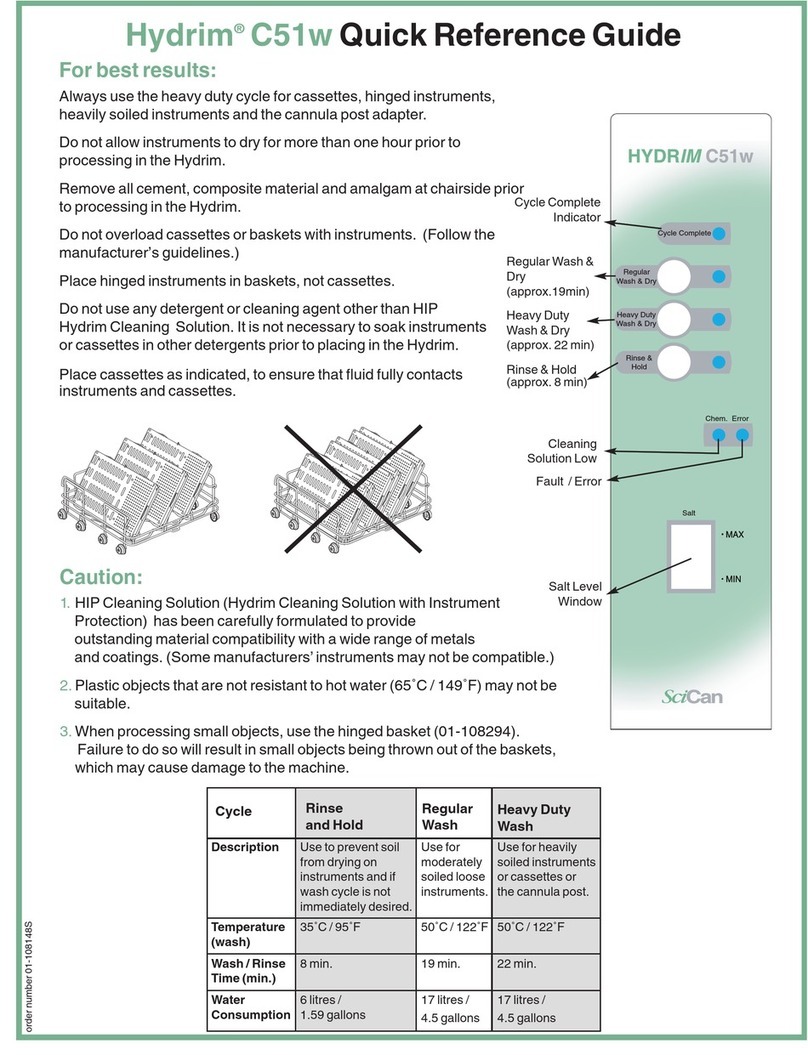
Promega MD1531 User manual

Revised 12/18
TM248
TECHNICAL MANUAL
Y Chromosome Deletion
Detection System,
Version 2.0
Instrucons for Use of Product MD1531
For Research Use Only. Not for use in diagnostic procedures.

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 1
www.promega.com TM248 · Revised 12/18
All technical literature is available at: www.promega.com/protocols/
Visit the web site to verify that you are using the most current version of this Technical Manual.
Y Chromosome Deletion
Detection System, Version 2.0
1. Description.........................................................................................................................................2
2. Product Components and Storage Conditions........................................................................................3
3. System Requirements ..........................................................................................................................3
3.A. Template....................................................................................................................................3
3.B. Thermal Cyclers .........................................................................................................................4
3.C. Contamination Control ............................................................................................................... 4
3.D. Control Reactions.......................................................................................................................5
4. Amplification Reactions.......................................................................................................................6
4.A. Reaction Setup for Large Sample Numbers ...................................................................................6
4.B. Thermal Cycling: Protocols for use with the Perkin-Elmer Model 480 and GeneAmp®PCR System
9600 or 9700 Thermal Cyclers.....................................................................................................8
4.C. Agarose Gel Electrophoresis ........................................................................................................9
4.D. Data Analysis—Controls............................................................................................................ 10
4.E. Data Analysis—Experimental Samples........................................................................................ 10
5. Troubleshooting................................................................................................................................ 12
6. References........................................................................................................................................ 14
7. Appendix.......................................................................................................................................... 15
7.A. Composition of Buffers and Solutions......................................................................................... 15
7.B. Related Products ...................................................................................................................... 15
7.C. Reaction Setup for Small Sample Numbers ................................................................................. 15
7.D. Preparation of Agarose Gels ...................................................................................................... 17
7.E. Worksheets.............................................................................................................................. 18
8. Summary of Changes ......................................................................................................................... 21

2Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
1. Description
The Y Chromosome Deletion Detection System, Version 2.0(a,b), is intended for research use only and provides a rapid
method for the detection of specific regions of the human Y chromosome. This system is designed to detect deletions
occurring in YqAZF. Further mapping of common deletion breakpoints relative to palindromes 1 through 8 may be
performed in additional experiments (1). This system consists of 20 primer pairs that are homologous to previously
identified and mapped sequence-tagged sites (STS; 2–6). This product is not intended for use in diagnostic
applications.
These primers will amplify nonpolymorphic short DNA segments from the Y chromosome when used in polymerase
chain reactions (PCR; 1). The primers have been combined into five sets for use in multiplex PCR. This makes it
possible to determine the presence or absence of all 20 sequence-tagged sites by performing five parallel PCR
amplifications.
Four of the Multiplex Master Mix sets (Multiplexes A–D) contain a control primer pair that amplifies a fragment of the
X-linked SMCX locus. The fifth Multiplex Master Mix set, Multiplex E, contains a control primer pair that amplifies a
unique region in both male and female DNA (ZFX/ZFY). These control primer pairs are internal controls for the
amplification reaction and the integrity of the genomic DNA sample. Finally, Multiplex E Master Mix includes a primer
pair that amplifies a region of the SRY gene.
Figure 1 shows a typical amplification of male genomic DNA, as well as a negative DNA control, for each of the five
Multiplex Master Mixes.
Figure 1. Multiplex analysis of wildtype male genomic DNA. Lane 1 shows the amplification products from
reactions using Multiplex A Master Mix with male DNA, and lane 2 shows the no-DNA control. Lane 3 shows the
amplification products from reactions using Multiplex B Master Mix with male DNA, and lane 4 shows the no-DNA
control. Lane 5 shows the amplification products from reactions using Multiplex C Master Mix with male DNA, and
lane 6 shows the no-DNA control. Lane 7 shows the amplification products from reactions using Multiplex D Master
Mix with male DNA, and lane 8 shows the no-DNA control. Lane 9 shows the amplification products from reactions
using Multiplex E Master Mix with male DNA, and lane 10 shows the no-DNA control. Lanes M contain the 50bp DNA
Step Ladder (Cat.# G4521). The reactions were performed on a Perkin-Elmer Model 480 thermal cycler. The gel is a
4% NuSieve®3:1 Plus TBE Buffer Reliant®gel.
4322TA09_3A
M
50 –
200 –
300 –
400 –
500 –
bp
100 –
– 50
– 200
– 300
– 400
– 500
– 100
A
12M
B
34M
C
56M
D
78M M
E
9 10

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 3
www.promega.com TM248 · Revised 12/18
2. Product Components and Storage Conditions
PRODUCT SIZE CAT.#
Y Chromosome Deleon Detecon System, Version 2.0 25 reacons MD1531
For Research Use Only. Not for use in diagnostic procedures. The Y Chromosome Deletion Detection System,
Version 2.0, includes:
• 500µl Multiplex A Master Mix
• 500µl Multiplex B Master Mix
• 500µl Multiplex C Master Mix
• 500µl Multiplex D Master Mix
• 500µl Multiplex E Master Mix
•200u GoTaq®DNA Polymerase
• 1ml Nuclease-Free Water
• 90µg 50bp DNA Step Ladder (340ng/µl)
• 2.5µg Male Genomic DNA (50ng/µl)
• 1ml Blue/Orange 6X Loading Dye
Storage Conditions: Store all components at –20°C. Avoid multiple freeze-thaw cycles. The Nuclease-Free Water
may be stored at room temperature (22–25°C).
3. System Requirements
3.A. Template
DNA concentration, purity and size are important considerations to ensure success with the Y Chromosome Deletion
Detection System, Version 2.0. DNA should be free of contaminating protein and salts. The DNA should not be
sheared. Poor-quality DNA may result in increased background or amplification failure. Adding too much or too little
DNA to the reaction can cause amplification failure. Failure can be manifested in several ways; sometimes complete
lack of amplification of all bands is observed with all master mixes, and in other cases dropout of all or portions of the
alleles in individual master mixes will be observed. Several commercially available DNA purification systems, including
the Wizard®Genomic DNA Purification Kit (Cat.# A1120, A1125, A1620) and the MagneSil®KF, Genomic System
(Cat.# MD1460), produce DNA of sufficient quality for the Y Chromosome Deletion Detection System.
Quantitation of DNA
We recommend quantitating the DNA prior to use in the Y Chromosome Deletion Detection System; either too much or
too little DNA can cause the reactions to fail. Absorbance readings at 260nm can be used to estimate DNA
concentration where 1Au = 50µg of double-stranded DNA/ml. Fifty nanograms of DNA should be used in each
amplification reaction.

4Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
3.A. Template (continued)
DNA Quality
High-quality DNA typically has an A260/A280 ratio of ≥1.8. The presence of impurities in the DNA sample can cause
amplification failure. We have observed that DNA concentration can be overestimated by spectrophotometry if the
A260/A280 ratio is low. When low amounts of DNA are added to the Y Chromosome Deletion Detection System, band
dropout in one or more of the master mixes can be observed. The amount of DNA in a sample can be verified by
ethidium bromide staining of DNA following electrophoresis in a 1% agarose gel. One hundred nanograms of genomic
DNA should be visible on an ethidium bromide-stained gel visualized by UV transillumination. The Male Genomic
DNA standard supplied in the Y Chromosome Deletion Detection System can be used as an electrophoresis standard.
Figure 2 is an example of an agarose gel containing ethidium bromide-stained genomic DNA samples of 100ng of DNA
as determined by A260. Differences in the intensity of the bands indicate different DNA concentrations that result from
variation in the A260/A280 ratio.
Figure 2. Gel image of 100ng (based on A260) genomic DNA of varying quality. The end lanes contain 100ng
of Male Genomic DNA supplied with the system.
3.B. Thermal Cyclers
The Y Chromosome Deletion Detection System has been optimized for use with either the Perkin-Elmer GeneAmp®
PCR System 9600 or 9700 or the Perkin-Elmer Model 480 thermal cycler. Use of other thermal cyclers may require
optimization of cycling parameters. We recommend validating with actual samples in addition to the positive control,
as the preparation of the sample DNA can affect the cycling conditions. The Perkin-Elmer Model 480 thermal cycler
program is a good starting point for thermal cyclers without programmed ramps, and the Perkin-Elmer GeneAmp®
PCR System 9700 cycling program is a good starting point for thermal cyclers with programmed ramps.
3.C. Contamination Control
Preventing DNA contamination of reaction components is essential. Always use aerosol-resistant pipette tips and clean
gloves, and avoid carryover contamination of stock solutions. Reagents that are used for amplification reactions should
be maintained and used separately from completed amplification reactions, preferably in separate pre-amplification
and post-amplification locations.

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 5
www.promega.com TM248 · Revised 12/18
3.D. Control Reactions
Suitable control reactions should be included in each experiment.
• The positive Male Genomic DNA control should always yield the expected amplification products (see Section
7.E, Table 1, for expected sizes).
• The negative no-DNA control reaction will verify that the reagents are not contaminated with DNA (Figure 1;
lanes 2, 4, 6, 8 and 10).
Figure 3. Schematic representation of the Y Chromosome Deletion Detection System assay.
Sample
DNA
Positive
Control Male
Genomic DNA
Negative
Control
No DNA
3856MA10_2A
Dilute sample DNA
(in Nuclease-Free Water),
and prepare controls
Prepare Multiplex Master Mix
and TaqDNA polymerase mixtures,
one for each Multiplex Master Mix.
Prepare reactions:
1. Place sample DNA
or control in tubes.
2. Add Multiplex
Master Mix and TaqDNA
polymerase mixture.
Thermal cycling
Agarose gel electrophoresis
A
A
B
C
D
E
BC D
Sample
DNA
Positive
Control
Negative
Control
E
TaqDNA
polymerase for
each sample
Multiplex
Master Mix for
each sample

6Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
4. Amplification Reactions
Materials to Be Supplied by the User
(Solution compositions are provided in Section 7.A.)
• nuclease-free light mineral oil
• Perkin-Elmer Model 480 or GeneAmp®PCR System 9600 or 9700 thermal cycler
• Amplification tubes compatible with thermal cycler. For the Perkin Elmer Model 480 thermal cycler, use 0.5ml
thin-walled GeneAmp®reaction tubes. For the GeneAmp®PCR System 9600 and 9700 thermal cyclers, use
0.2ml thin-walled MicroAmp®reaction tubes or a MicroAmp®optical 96-well reaction plate.
• 4% NuSieve®3:1 Plus TBE Buffer agarose precast Reliant®minigels (Cambrex
• Cat.# 54927, 54928 or 54929), Latitude®precast minigels agarose (Cambrex
• Cat.# 57222 or 57232), or NuSieve®3:1 agarose (Cambrex Cat.# 50090 or 50091)
• 1X TBE buffer
• ethidium bromide
• aerosol-resistant pipette tips
• UV transilluminator
Caution: Ethidium bromide is a suspected carcinogen. Always wear gloves when working with ethidium bromide
solutions.
4.A. Reaction Setup for Large Sample Numbers
This protocol is designed to analyze multiple samples at one time. If you are analyzing only 1 or 2 samples, see
Section 7.C.
In this procedure:
• Aliquots of the sample DNAs are placed in the reaction tube.
• Taq DNA polymerase is added to each of the Multiplex Master Mixes.
• The Multiplex Master Mixes containing Taq DNA polymerase are added to the reaction tubes. Each sample is
analyzed with all five Multiplex Master Mixes.
Protocol
1. Thaw the Multiplex Master Mixes. Vortex for 5–10 seconds, and store on ice. Thaw the Nuclease-Free Water and
Male Genomic DNA.
2. Determine the number of reactions for each Multiplex Master Mix. For each sample there will be five reaction
tubes, one for each Multiplex Master Mix. Include a positive Male Genomic DNA control and a negative no-DNA
control for each Multiplex Master Mix.
!

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 7
www.promega.com TM248 · Revised 12/18
Multiplex
Master Mix
# of Sample
DNAs
Positive Control
(Male Genomic
DNA)
Negative Control
(No DNA)
Total Reaction
Tubes
A 1 1
B 1 1
C 1 1
D 1 1
E 1 1
3. Set up and label the required number of reaction tubes as determined above. Use thin-walled amplification tubes
or an optical plate. Place on ice.
4. In a separate tube, dilute each sample DNA to 10ng/µl using the supplied Nuclease-Free Water. Mix well by
vortexing for 5–10 seconds. Add 5µl of diluted DNA to the appropriately labeled reaction tubes (above) on ice.
Dilute the Male Genomic DNA to 10ng/µl by adding 6µl of Male Genomic DNA to 24µl of Nuclease-Free Water.
Mix well by vortexing for 5–10 seconds. Add 5µl to appropriately labeled reaction tubes (above) on ice.
For the negative control (no DNA), add 5µl of Nuclease-Free Water to the appropriately labeled reaction tubes
(above) on ice.
5. Prepare five Multiplex Master Mix and Taq DNA polymerase mixtures on ice, one for each Multiplex Master Mix.
Vortex the Multiplex Master Mixes before using them.
Component
Volume per
Reaction
Volume per
10 Reactions
Multiplex Master Mix 20µl 200µl
Taq DNA Polymerase (5u/µl) 0.2µl 2µl
Final Volume 20.2µl 202µl
6. Vortex to mix.
7. Add 20µl of the Multiplex Master Mix and Taq DNA polymerase mixture to the appropriate reaction tubes,
which contain the sample DNA or controls, on ice.
8. Gently vortex to mix.
9. Centrifuge briefly to bring the contents to the bottom of the tubes. Place on ice until ready for thermal cycling.
10. If using a thermal cycler that requires mineral oil, tilt the tubes and add one drop of oil to the side of the tube,
letting the oil run down the side of the tube.

8Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
4.B. Thermal Cycling: Protocols for use with the Perkin-Elmer Model 480 and GeneAmp®PCR System
9600 or 9700 Thermal Cyclers
Place the tubes in the thermal cycler, and run the recommended program. The preferred protocols for use with the
Perkin-Elmer Model 480 and GeneAmp®PCR System 9600 and 9700 thermal cyclers are provided below. It may be
necessary to optimize the program with other thermal cyclers.
Note: It is important to preheat the instrument to 94°C before placing tubes into the machine.
Program for the Perkin-Elmer Model 480 Thermal Cycler
Preheat the thermal cycler to 94°C before placing tubes inside.
Cycling Profile:
94°C for 2 minutes, then:
94° for 1 minute
57°C for 30 seconds
72°C for 1 minute
For 35 cycles, then:
72°C for 5 minutes
4°C soak
After completion of the thermal cycling protocol, store the samples at –20°C.
Program for the Perkin-Elmer GeneAmp®PCR System 9600 Thermal Cycler
Preheat the thermal cycler to 94°C before placing tubes inside.
Cycling Profile:
94°C for 2 minutes, then:
94°C for 30 seconds
Ramp 68 seconds to 58°C, hold 30 seconds
Ramp 50 seconds to 70°C, hold 45 seconds
For 35 cycles, then:
68°C for 2 minutes
4°C soak
After completion of thermal cycling protocol, store samples at –20°C.
!

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 9
www.promega.com TM248 · Revised 12/18
Program for the Perkin-Elmer GeneAmp®PCR System 9700 Thermal Cycler
Preheat the thermal cycler to 94°C before placing tubes inside.
Cycling Profile:
94°C for 2 minutes, then,
94°C for 30 seconds
30% Ramp to 58°C, hold 30 seconds
30% Ramp to 70°C, hold 45 seconds
Repeat for 35 cycles, then:
68°C for 2 minutes
4°C soak
(9600 default mode)
After completion of the thermal cycling protocol, store samples at –20°C.
4.C. Agarose Gel Electrophoresis
1. For optimal visualization of the amplification products we recommend using a 4% NuSieve®3:1 Plus TBE buffer
precast gel. Alternatively, cast a 4% NuSieve®3:1 agarose gel in 1X TBE buffer containing 0.5µg/ml ethidium
bromide. For instructions for the preparation of agarose gels, see Section 7.D.
Note: The running buffer should contain 0.5µg/ml ethidium bromide.
2. Dilute the molecular weight marker as follows:
Volume
50bp DNA Step Ladder 12µl
Blue/Orange 6X Loading Dye 4µl
Nuclease-Free Water 8µl
3. Add 2.5µl of the Blue/Orange 6X Loading Dye to each amplification tube, and mix.
4. Load 5–10µl of each sample and 10µl of the diluted molecular weight markers onto the gel.
5. Run the gel in 1X TBE buffer containing 0.5µg/ml ethidium bromide at 5V/cm (measured as the distance
between the electrodes) until the bromophenol blue dye front migrates to the bottom of the gel.
6. Photograph the gel using a UV transilluminator (320nm).
!

10 Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
4.D. Data Analysis—Controls
Determine that the control reactions produced the expected results before analyzing your samples. The worksheets
provided (see Section 7.E, Table 1) can be used for analysis of both the control and experimental samples.
1. Negative No-DNA Control: There should be no specific amplification products in the lanes containing the
negative no-DNA control reactions. There may be some low-molecular-weight bands or smearing that are the
result of primer interactions. Amplification products in the negative no-DNA control reactions are indicative of
contaminating DNA, and the experiment should be repeated with care to avoid contamination.
2. Positive Male Genomic DNA Control: The number and size of the amplification products for each Multiplex
Master Mix are indicated in Table 1 (Section 7.E). The positive Male Genomic DNA control reaction for each
Multiplex Master Mix should have all the bands indicated for that Multiplex Master Mix (Table 1). The sizes of
the amplification products can be estimated by comparison with the DNA markers. If all of the expected bands
are not present in the reactions with positive Male Genomic DNA control, or if there are prominent extra bands,
it is indicative of a problem with the amplification reagents or the thermal cycler (see Section 5). Results should
not be considered valid if any of the expected amplification products are missing in the reactions with the positive
Male Genomic DNA control.
3. Control Primer in Multiplex Master Mixes: In Multiplexes A, B, C, and D reactions the smallest
amplification product (83bp) should be produced from an X-linked locus (SMCX). In Multiplex E the largest
(496bp) amplification product should be produced from the ZFY/ZFX genes. The absence of these products is
indicative of a problem with that particular PCR amplification. If the control bands are present with the Male
Genomic DNA control but not with the sample DNA, it suggests that there may be a problem with the genomic
DNA used as a template, such as the presence of impurities, inaccurate DNA quantitation or degraded DNA.
Check the DNA template on an agarose gel before repeating the amplification. Repeat any reactions in which
the control product is absent. It may be necessary to isolate the template DNA again. We recommend using
the Wizard®Genomic DNA Purification Kit (Section 7.B; Cat.# A1120, A1125, A1620) or the MagneSil®KF,
Genomic System (Cat.# MD1460) for isolation of template DNA. Ideally, DNA should be eluted in water.
4. Female DNA Control (optional): Some researchers include a female DNA control. For Multiplexes A, B, C
and D, the smallest product (83bp) and the largest product (496bp) for Multiplex E should be present when
female DNA is used as a template. Nonspecific bands may appear when female genomic DNA is used. These
bands are generally faint and do not correspond in size to male-specific bands. With Multiplex E, a distinct high-
molecular-weight band is generally seen.
4.E. Data Analysis—Experimental Samples
The worksheets provided in the Appendix, Section 7.E, can be used for analysis of experimental samples. Determine
the presence or absence of the expected PCR products (Table 1). If there are any products absent from the reactions,
they can be mapped using the Y Chromosome Map Worksheet (Table 2). All deletions should be contiguous. Figure 4
shows an example of gel analysis of DNA carrying a Y chromosome deletion.
Adjacent regions of the Y chromosome do not appear as sequential amplification products using the Y Chromosome
Deletion Detection System. Exceptions are SY242 and SY208 of the DAZ locus, which are represented as sequential
amplification products in Multiplex B, and SY84 and SY86 of the DYS273 and DYS148 loci, which are represented as
sequential amplification products in Multiplex E.

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 11
www.promega.com TM248 · Revised 12/18
If multiple amplification bands are missing, and those bands do not map to adjacent regions of the Y chromosome
(Table 2), they represent dropout bands and are not deletions. See the troubleshooting guide (Section 5) for help if you
observe dropout of alleles in individual master mixes.
We have observed some nonspecific bands that appear above or below the expected amplification products on the
agarose gel. If your negative controls show no detectable PCR products, these nonspecific bands should have no effect
on analysis.
Figure 4. Y chromosome deletion analysis. The amplification products from Multiplex A Master Mix, Multiplex B
Master Mix, Multiplex C Master Mix, Multiplex D Master Mix and Multiplex E Master Mix reactions are shown. Lanes:
1, Male Genomic DNA control; 2, sample male DNA (containing deletions). Bands generated with Male Genomic DNA
control are compared to those generated with a sample containing Y chromosome deletions. (Note the deleted bands in
Multiplex A–D. Multiplex E shows no deletions.) The marker (M) lanes contain the 50bp DNA Step Ladder
(Cat.# G4521).
M12M12 M12
4389TA11_3A
A. B. C.
M1 2M
12
D. E.

12 Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
5. Troubleshooting
For questions not addressed here, please contact your local Promega Branch Office or Distributor. Contact information
Symptoms Causes and Comments
Low yield or no Thermal cycler programmed incorrectly. Verify that the times
amplification product and temperatures are correct.
Thermal cycler not properly calibrated. Perform a set of
control reactions to determine if certain positions in the
thermal cycler yield little or no product.
Top of thermal cycler open. The top must be closed for correct
heating and cooling.
Missing reaction component. Check the reaction components,
and repeat the reaction.
Degraded reagent. Store Multiplex Master Mixes at –20°C.
Keep on ice once thawed. Avoid multiple freeze-thaw cycles.
Incorrect tubes. Use only thin-walled microcentrifuge tubes
for amplification. For the Perkin-Elmer Model 480 thermal
cycler, use 0.5ml thin-walled GeneAmp®reaction tubes, and
for the GeneAmp®PCR System 9600 or 9700 thermal cycler,
use 0.2ml thin-walled MicroAmp®reaction tubes or
MicroAmp®optical 96-well reaction plate.
Inhibitors in the DNA sample. DNA should be free of
contaminating proteins and salts. Repurify or ethanol
precipitate the DNA sample to remove inhibitors.
Too much or too little DNA used. DNA should be quantitated
prior to use in the Y Chromosome Deletion Detection System.
Use 50ng of high-molecular-weight (nonsheared) DNA for
each amplification reaction or 250ng total for each sample.
Inaccurate quantitation of DNA. DNA concentration can be
overestimated by spectrophotometry if the A260/A280 ratio is
low. Verify DNA concentration and quality by ethidium
bromide staining after electrophoresis on a low percent
(1–1.2%) agarose gel. We recommend comparing sample
DNA with genomic DNA of known concentration such as the
Male Genomic DNA supplied with the system. One hundred
nanograms of the Male Genomic DNA is clearly visible on
ethidium bromide-stained gels.
Poor mixing of components. Concentration gradients can
form in Multiplex Master Mixes stored at –20°C. Mix
thoroughly by vortexing for 10–15 seconds before use.

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 13
www.promega.com TM248 · Revised 12/18
Symptoms Causes and Comments
Low yield or no The thermal cycler was not preheated prior to placing tubes
amplification product (continued) inside. Preheat the instrument to 94°C.
Multiple nonspecific Thermal cycler programmed incorrectly. Verify that the times
amplification products and temperatures are correct.
Reactions were not set up on ice. Make sure to set up reactions
on ice.
Too much genomic DNA. We recommend using 50ng of DNA
per reaction.
Too much Taq DNA polymerase. Do not use more than 1 unit
of Taq DNA polymerase per reaction.
Annealing temperature too low. Check the accuracy of the
thermal cycler. Increase the annealing temperature in
0.5–1°C increments.
Missing bands in control reaction Annealing temperature too high. Check the accuracy of the
thermal cycler. Decrease the annealing temperature in 0.5–1°C
increments.
Internal control band is missing in Sample DNA degraded or of poor quality. Repeat reactions.
sample DNA but present in control You may need to re-isolate sample DNA. We recommend the
Wizard®Genomic DNA Purification Kit or MagneSil®KF,
Genomic System.
Dropout of bands in Too little DNA used. DNA should be quantitated prior to use
amplification of sample DNA in the Y Chromosome Deletion Detection System. Use 50ng
for each amplification reaction or 250ng total for each sample.
DNA concentration can be overestimated by spectrophotometry
if the A260/A280 ratio is low. Verify DNA concentration by
ethidium bromide staining after electrophoresis on a low
percent (1–1.2%) agarose gel. We recommend comparing
sample DNA with genomic DNA of known concentration
such as the Male Genomic DNA supplied with the system.
One hundred nanograms of the Male Genomic DNA is clearly
visible on ethidium bromide-stained gels.
Poor DNA purity. High-quality DNA typically has an
A260/A280 ratio of ≥1.8. However, A260/A280 ratio may not
accurately predict purity. Impurities in the sample may cause
amplification failure. To test for impurities, add some of your
sample DNA to the positive control. Subsequent inhibition of
this “spiked” positive control reaction indicates that
impurities are in the sample DNA.

14 Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
5. Troubleshooting (continued)
Symptoms Causes and Comments
Dropout of bands in Suboptimal conditions. The system is tested using the Perkin-
amplification of sample DNA (continued) Elmer GeneAmp®PCR System 9600, 9700 and the Model 480
thermal cyclers. If you are using a different thermal cycler,
you will need to optimize amplification conditions.
EDTA in sample. EDTA at a concentration of ≥0.4mM can
adversely affect amplification.
Incorrect protocol. The protocol must be followed precisely.
Wrong reaction volume. Standard reactions are 25µl.
Reactions of 12.5µl are prone to dropout of multiple bands.
Bands in negative control reactions Cross-contamination. Pre- and post-amplification areas
should be isolated, preferably in separate rooms. Use aerosol-
resistant tips and dedicated pipettes for assembling reactions.
Workstations and pipettes can be cleaned with a mild bleach
solution before and after use.
6. References
1. Skaletsky, H. et al. (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence
classes. Nature 423, 825–37.
* Especially note Figure 2, available online as data supplementary to the article.
2. Vollrath, D. et al. (1992) The human Y chromosome: A 43-interval map based on naturally occurring deletions.
Science 258, 52–9.
3. Foote, S. et al. (1992) The human Y chromosome: Overlapping DNA clones spanning the euchromatic region.
Science 258, 60–6.
4. Affara, N. et al. (1996) Report of the second international workshop on Y chromosome mapping 1995.
Cytogenet. Cell. Genet. 73, 33–76.
5. Kent-First, M.G. et al. (1996) Gene sequence and evolutionary conservation of human SMCY. Nat. Genet. 14,
128–9.
6. Vogt, P.H. et al. (1997) Report of the Third International Workshop on Y-chromosome mapping 1997.
Cytogenet. Cell Genet. 79, 1–20.
7. Kostiner, D.R., Turek, P.K. and Reijo, R.A. (1998) Male infertility: Analysis of the markers and genes on the
human Y chromosome. Hum. Reprod. 13, 3032–8.
8. Lahn, B.T. and Page, D.C. (1997) Functional coherence of the human Y chromosome. Science 278, 675–80.
9. Reijo, R. et al. (1996) Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y
chromosome. Lancet 347, 1290–3.

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 15
www.promega.com TM248 · Revised 12/18
10. Repping, S. et al. (2002) Recombination between palindromes P5 and P1 on the human Y chromosome causes
massive deletions and spermatogenic failure. Am. J. Hum. Genet. 71, 906–22.
11. Saxena, R. et al. (1996) The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that
was transposed, repeatedly amplified and pruned. Nat. Genet. 14, 292–9.
7. Appendix
7.A. Composition of Buffers and Solutions
1X TBE buffer
89mM Tris-base
110mM boric acid
2mM EDTA
7.B. Related Products
Accessory Components
Product Size Cat.#
Nuclease-Free Water (2 × 25ml) 50ml P1193
Mineral Oil 12ml DY1151
DNA Purification Systems
Product Size Cat.#
Wizard®Genomic DNA Purification Kit 100 isolations × 300µl A1120
500 isolations × 300µl A1125
100 isolations × 10ml A1620
MagneSil®KF, Genomic System 200 preps MD1460
7.C. Reaction Setup for Small Sample Numbers
This protocol is designed to analyze a small number of samples. The order of addition of Taq DNA polymerase,
Multiplex Master Mixes and genomic DNA is different from the Large Sample Number Protocol (Section 4.A). These
changes conserve the Multiplex Master Mixes to ensure the full number of reactions can be performed.
In this procedure:
• The Multiplex Master Mixes are added to the reaction tubes.
• A mixture of genomic DNA and Taq DNA polymerase in Nuclease-Free Water is prepared.
• The genomic DNA/Taq DNA polymerase mixture is added to each Multiplex Master Mix.
• Each sample DNA is analyzed with all five of the Multiplex Master Mixes.

16 Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
7.C. Reaction Setup for Small Sample Numbers (continued)
An alternative protocol for large sample numbers is also provided (Section 4.A).
1. Thaw the Multiplex Master Mixes. Vortex for 5–10 seconds, and store on ice. Thaw the Nuclease-Free Water and
Male Genomic DNA.
2. Determine the number of reactions to perform for each Multiplex Master Mix. For each sample there will be five
reaction tubes, one for each Multiplex Master Mix. Include a positive Male Genomic DNA control and negative
no-DNA control for each Multiplex Master Mix.
Multiplex
Master Mix
# of Sample
DNAs
Positive Control
(Male Genomic
DNA)
Negative Control
(No DNA)
Total Reaction
Tubes
A 1 1
B 1 1
C 1 1
D 1 1
E 1 1
3. Label the required number of reaction tubes as determined above. For the Perkin-Elmer Model 480 thermal
cycler, use 0.5ml thin-walled GeneAmp®reaction tubes, and for the GeneAmp®PCR System 9600 or 9700
thermal cycler, use 0.2ml thin-walled MicroAmp®reaction tubes or MicroAmp®optical 96-well reaction plate.
Place the reaction tubes on ice.
4. Vortex the Multiplex Master Mixes for 5–10 seconds. Place 20µl of each Multiplex Master Mix in the
appropriately labeled reaction tubes on ice.
5. For each DNA sample, prepare a genomic DNA mixture on ice following the table below.
Component
Single
Reactions
Duplicate
Reactions
Sample DNA ~250ng ~500ng
Taq DNA polymerase (5u/µl) 1µl 2µl
Nuclease-Free Water to a final volume of 25µl 50µl
For the positive Male Genomic DNA control reactions, combine 5µl of Male Genomic DNA, 1µl of Taq DNA
polymerase (5u/µl) and 19µl of Nuclease-Free Water on ice. Vortex to mix.
For the negative no-DNA control, add 1µl Taq DNA polymerase (5u/µl) to 25µl of Nuclease-Free Water on ice.
Vortex gently to mix.
6. Add 5µl genomic DNA or control mixture prepared in Step 5 to the appropriate reaction tubes containing the
Multiplex Master Mixes on ice. Vortex to mix.

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 17
www.promega.com TM248 · Revised 12/18
7. Centrifuge briefly to bring the contents to the bottom of the tubes. Place reaction tubes on ice until ready for
thermal cycling.
8. If using a thermal cycler that requires mineral oil, tilt the tubes and add one drop of oil to the side of the tube,
letting the oil run down the side of the tube.
7.D. Preparation of Agarose Gels
1. Use NuSieve®3:1 agarose available from Cambrex.
2. Choose a beaker 2–4 times the volume of the agarose solution.
3. Add the appropriate amount of NuSieve®3:1 agarose and 1X TBE buffer to make a 4% gel (i.e., 100ml of 1X
TBE buffer and 4g NuSieve®3:1 agarose) to the beaker.
4. Weigh the beaker, or mark the level of the liquid in the beaker.
5. Mix to wet agarose.
6. Soak the agarose for 15 minutes at room temperature.
7. Heat the beaker in a microwave until the solution boils.
8. Boil the agarose for 1 minute or until it is completely dissolved.
9. Remove the agarose from the microwave.
10. Gently swirl the beaker to mix.
Caution: Any microwaved solution may become superheated and boil over when agitated.
11. Add sufficient hot distilled water to obtain the initial weight or volume.
12. Mix thoroughly.
13. Cool the solution to 50–60°C.
14. Add ethidium bromide to 0.5µg/ml.
15. Pour the gel to a depth of 5mm. It is important that the gel is thin for optimum resolution.
16. Allow the gel to set.
17. Run gel in 1X TBE buffer containing 0.5µg/ml ethidium bromide. For best resolution, it is important ethidium
bromide is in both the running buffer and the gel.
18. Load samples.
19. Run gel at 5V/cm until the bromophenol blue dye front migrates to the bottom of the gel.
!
18 Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516
TM248 · Revised 12/18 www.promega.com
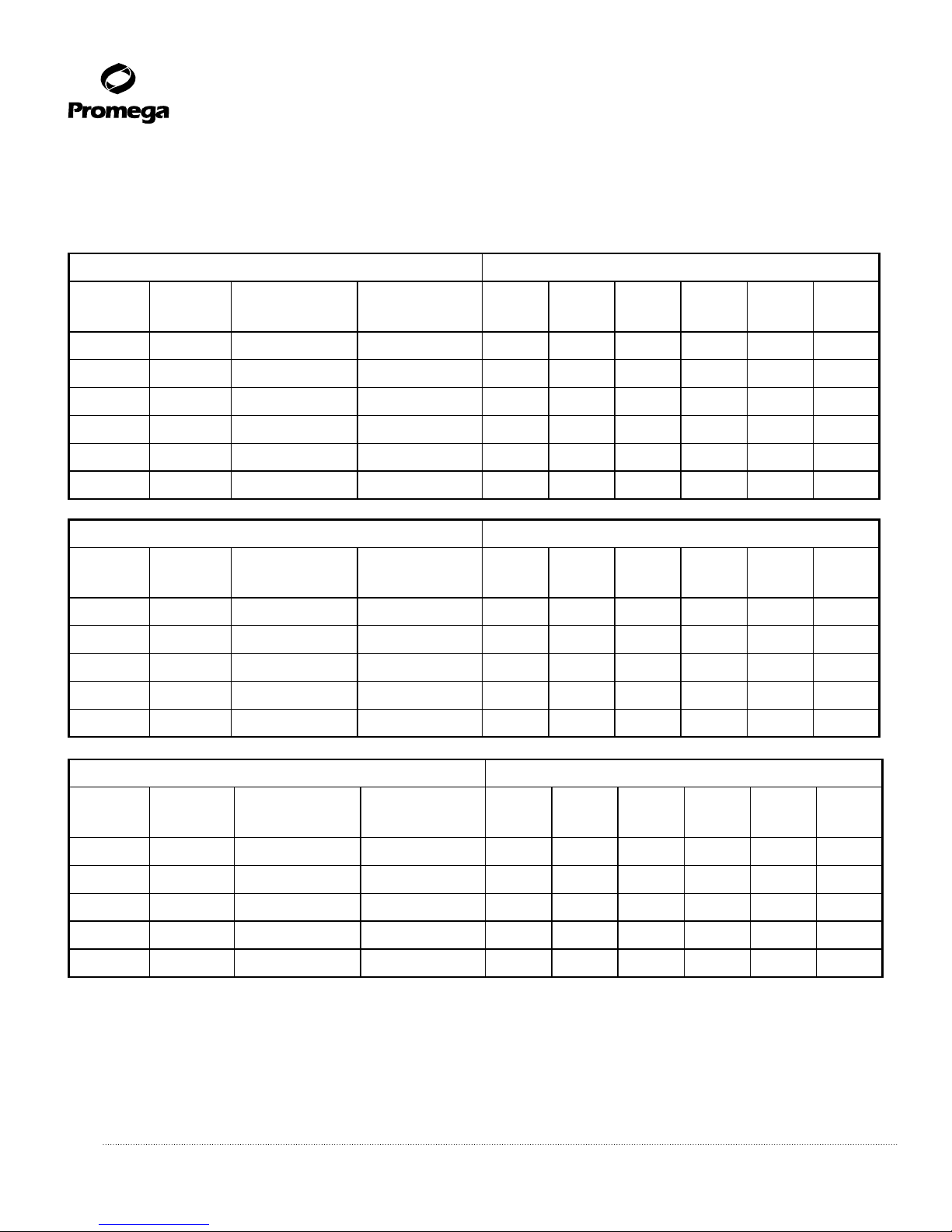
7.E. Worksheets
Table 1. PCR Amplification Product Profile for Test Samples.
Record the presence (+) or absence (–) of the PCR amplification products for each experimental sample.
Multiplex A Master Mix Samples (+/–)
STS
Locus
Size of
Product (bp)
Map
Position
SY254 DAZ 380 18
SY157 DYS240 290 20
SY81 DYS271 209 2
SY130 DYS221 173 11
SY182 KAL-Y 125 5
SMCX 83 Control
Multiplex B Master Mix Samples (+/–)
STS
Locus
Size of
Product (bp)
Map
Position
SYPR3 SMCY 362 7
SY127 DYS218 274 9
SY242 DAZ 233 16
SY208 DAZ 140 17
SMCX 83 Control
Multiplex C Master Mix Samples (+/–)
STS
Locus
Size of
Product (bp)
Map
Position
SY128 DYS219 228 10
SY121 DYS212 190 6
SY145
DYF51S1
143 14
SY255 DAZ 124 19
SMCX 83 Control

Promega Corporaon · 2800 Woods Hollow Road · Madison, WI 53711-5399 USA · Toll Free in USA 800-356-9526 · 608-274-4330 · Fax 608-277-2516 19
www.promega.com TM248 · Revised 12/18
Multiplex D Master Mix Samples (+/–)
STS
Locus
Size of
Product (bp)
Map
Position
SY133 DYS223 177 12
SY152 DYS236 125 15
SY124 DYS215 109 8
SMCX 83 Control
Multiplex E Master Mix Samples (+/–)
STS
Locus
Size of
Product (bp)
Map
Position
ZFX/ZFY 496 Control
SY14 SRY 400 1
SY134 DYS224 303 13
SY86 DYS148 232 3
SY84 DYS273 177 4
Table of contents
Other Promega Medical Equipment manuals